1kcf: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
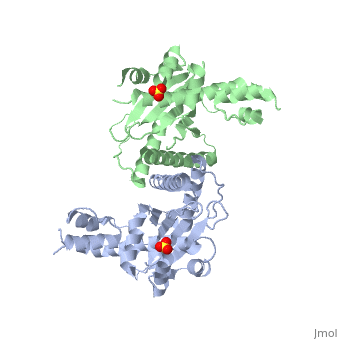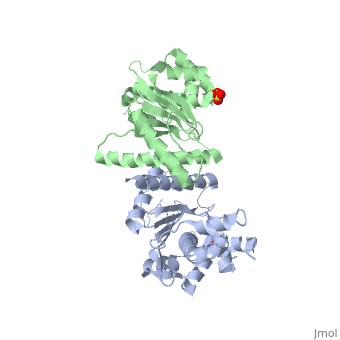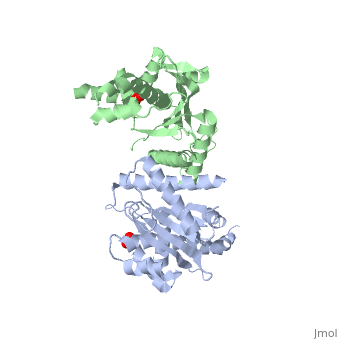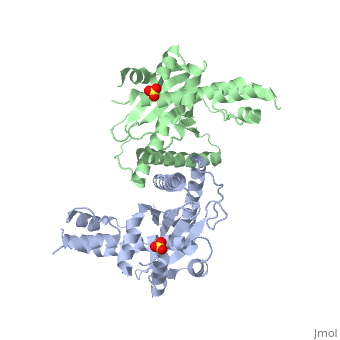
[[Image: | ==Crystal Structure of the Yeast Mitochondrial Holliday Junction Resolvase, Ydc2== | ||
<StructureSection load='1kcf' size='340' side='right' caption='[[1kcf]], [[Resolution|resolution]] 2.30Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[1kcf]] is a 2 chain structure with sequence from [http://en.wikipedia.org/wiki/Schizosaccharomyces_pombe Schizosaccharomyces pombe]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1KCF OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1KCF FirstGlance]. <br> | |||
</td></tr><tr><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene><br> | |||
<tr><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">SPAC25G10.02 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=4896 Schizosaccharomyces pombe])</td></tr> | |||
<tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1kcf FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1kcf OCA], [http://www.rcsb.org/pdb/explore.do?structureId=1kcf RCSB], [http://www.ebi.ac.uk/pdbsum/1kcf PDBsum]</span></td></tr> | |||
<table> | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/kc/1kcf_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/chain_selection.php?pdb_ID=2ata ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
Resolution of Holliday junctions into separate DNA duplexes requires enzymatic cleavage of an equivalent strand from each contributing duplex at or close to the point of strand exchange. Diverse Holliday junction-resolving enzymes have been identified in bacteria, bacteriophages, archaea and pox viruses, but the only eukaryotic examples identified so far are those from fungal mitochondria. We have now determined the crystal structure of Ydc2 (also known as SpCce1), a Holliday junction resolvase from the fission yeast Schizosaccharomyces pombe that is involved in the maintenance of mitochondrial DNA. This first structure of a eukaryotic Holliday junction resolvase confirms a distant evolutionary relationship to the bacterial RuvC family, but reveals structural features which are unique to the eukaryotic enzymes. Detailed analysis of the dimeric structure suggests mechanisms for junction isomerization and communication between the two active sites, and together with site-directed mutagenesis identifies residues involved in catalysis. | |||
Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2.,Ceschini S, Keeley A, McAlister MS, Oram M, Phelan J, Pearl LH, Tsaneva IR, Barrett TE EMBO J. 2001 Dec 3;20(23):6601-11. PMID:11726496<ref>PMID:11726496</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
==See Also== | ==See Also== | ||
*[[Resolvase|Resolvase]] | *[[Resolvase|Resolvase]] | ||
== References == | |||
== | <references/> | ||
< | __TOC__ | ||
</StructureSection> | |||
[[Category: Schizosaccharomyces pombe]] | [[Category: Schizosaccharomyces pombe]] | ||
[[Category: Barrett, T E.]] | [[Category: Barrett, T E.]] | ||
Revision as of 13:44, 28 September 2014
Crystal Structure of the Yeast Mitochondrial Holliday Junction Resolvase, Ydc2Crystal Structure of the Yeast Mitochondrial Holliday Junction Resolvase, Ydc2
Structural highlights
Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedResolution of Holliday junctions into separate DNA duplexes requires enzymatic cleavage of an equivalent strand from each contributing duplex at or close to the point of strand exchange. Diverse Holliday junction-resolving enzymes have been identified in bacteria, bacteriophages, archaea and pox viruses, but the only eukaryotic examples identified so far are those from fungal mitochondria. We have now determined the crystal structure of Ydc2 (also known as SpCce1), a Holliday junction resolvase from the fission yeast Schizosaccharomyces pombe that is involved in the maintenance of mitochondrial DNA. This first structure of a eukaryotic Holliday junction resolvase confirms a distant evolutionary relationship to the bacterial RuvC family, but reveals structural features which are unique to the eukaryotic enzymes. Detailed analysis of the dimeric structure suggests mechanisms for junction isomerization and communication between the two active sites, and together with site-directed mutagenesis identifies residues involved in catalysis. Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2.,Ceschini S, Keeley A, McAlister MS, Oram M, Phelan J, Pearl LH, Tsaneva IR, Barrett TE EMBO J. 2001 Dec 3;20(23):6601-11. PMID:11726496[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||