1dfu: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
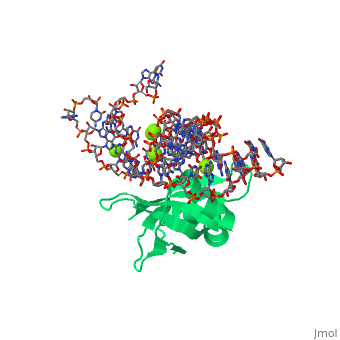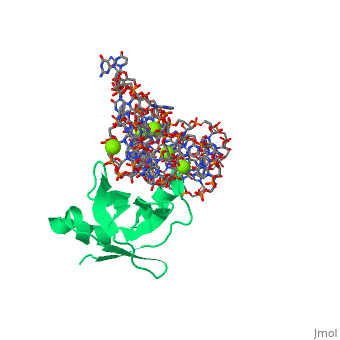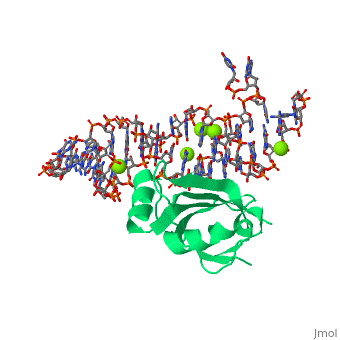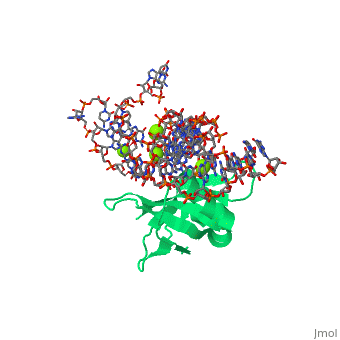
[[Image: | ==CRYSTAL STRUCTURE OF E.COLI RIBOSOMAL PROTEIN L25 COMPLEXED WITH A 5S RRNA FRAGMENT AT 1.8 A RESOLUTION== | ||
<StructureSection load='1dfu' size='340' side='right' caption='[[1dfu]], [[Resolution|resolution]] 1.80Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[1dfu]] is a 3 chain structure with sequence from [http://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1DFU OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1DFU FirstGlance]. <br> | |||
</td></tr><tr><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=MG:MAGNESIUM+ION'>MG</scene><br> | |||
<tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1dfu FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1dfu OCA], [http://www.rcsb.org/pdb/explore.do?structureId=1dfu RCSB], [http://www.ebi.ac.uk/pdbsum/1dfu PDBsum]</span></td></tr> | |||
<table> | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/df/1dfu_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/chain_selection.php?pdb_ID=2ata ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
The crystal structure of Escherichia coli ribosomal protein L25 bound to an 18-base pair portion of 5S ribosomal RNA, which contains "loop E," has been determined at 1.8-A resolution. The protein primarily recognizes a unique RNA shape, although five side chains make direct or water-mediated interactions with bases. Three beta-strands lie in the widened minor groove of loop E formed by noncanonical base pairs and cross-strand purine stacks, and an alpha-helix interacts in an adjacent widened major groove. The structure of loop E is largely the same as that of uncomplexed RNA (rms deviation of 0.4 A for 11 base pairs), and 3 Mg(2+) ions that stabilize the noncanonical base pairs lie in the same or similar locations in both structures. Perhaps surprisingly, those residues interacting with the RNA backbone are the most conserved among known L25 sequences, whereas those interacting with the bases are not. | |||
Structure of Escherichia coli ribosomal protein L25 complexed with a 5S rRNA fragment at 1.8-A resolution.,Lu M, Steitz TA Proc Natl Acad Sci U S A. 2000 Feb 29;97(5):2023-8. PMID:10696113<ref>PMID:10696113</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
==See Also== | ==See Also== | ||
*[[Ribosomal protein L25|Ribosomal protein L25]] | *[[Ribosomal protein L25|Ribosomal protein L25]] | ||
== References == | |||
== | <references/> | ||
< | __TOC__ | ||
</StructureSection> | |||
[[Category: Escherichia coli]] | [[Category: Escherichia coli]] | ||
[[Category: Lu, M.]] | [[Category: Lu, M.]] | ||
Revision as of 09:49, 4 September 2014
CRYSTAL STRUCTURE OF E.COLI RIBOSOMAL PROTEIN L25 COMPLEXED WITH A 5S RRNA FRAGMENT AT 1.8 A RESOLUTIONCRYSTAL STRUCTURE OF E.COLI RIBOSOMAL PROTEIN L25 COMPLEXED WITH A 5S RRNA FRAGMENT AT 1.8 A RESOLUTION
Structural highlights
Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedThe crystal structure of Escherichia coli ribosomal protein L25 bound to an 18-base pair portion of 5S ribosomal RNA, which contains "loop E," has been determined at 1.8-A resolution. The protein primarily recognizes a unique RNA shape, although five side chains make direct or water-mediated interactions with bases. Three beta-strands lie in the widened minor groove of loop E formed by noncanonical base pairs and cross-strand purine stacks, and an alpha-helix interacts in an adjacent widened major groove. The structure of loop E is largely the same as that of uncomplexed RNA (rms deviation of 0.4 A for 11 base pairs), and 3 Mg(2+) ions that stabilize the noncanonical base pairs lie in the same or similar locations in both structures. Perhaps surprisingly, those residues interacting with the RNA backbone are the most conserved among known L25 sequences, whereas those interacting with the bases are not. Structure of Escherichia coli ribosomal protein L25 complexed with a 5S rRNA fragment at 1.8-A resolution.,Lu M, Steitz TA Proc Natl Acad Sci U S A. 2000 Feb 29;97(5):2023-8. PMID:10696113[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences |
| ||||||||||||||||