Tropomyosin: Difference between revisions
Michal Harel (talk | contribs) No edit summary |
Michal Harel (talk | contribs) No edit summary |
||
| Line 57: | Line 57: | ||
[[2efr]], [[2efs]], [[2d3e]] - raTPM alpha-1 C-terminal+GNC4 leucine zipper – rabbit<br /> | [[2efr]], [[2efs]], [[2d3e]] - raTPM alpha-1 C-terminal+GNC4 leucine zipper – rabbit<br /> | ||
[[4a7f]], [[4a7h]], [[4a7l]] - raTPM α-1 + myosin + actin – Cryo EM<br /> | [[4a7f]], [[4a7h]], [[4a7l]] - raTPM α-1 + myosin + actin – Cryo EM<br /> | ||
[[3j4k]] - raTPM + actin – Cryo EM<br /> | |||
[[1kql]] - TPM alpha-1 C-terminal+GNC4 leucine zipper - rat<br /> | [[1kql]] - TPM alpha-1 C-terminal+GNC4 leucine zipper - rat<br /> | ||
[[1mv4]] - rTPM alpha-1 C-terminal <br /> | [[1mv4]] - rTPM alpha-1 C-terminal <br /> | ||
| Line 68: | Line 69: | ||
==References== | ==References== | ||
<references /> | <references /> | ||
[[Category: Topic Page]] | |||
Revision as of 10:07, 25 May 2014
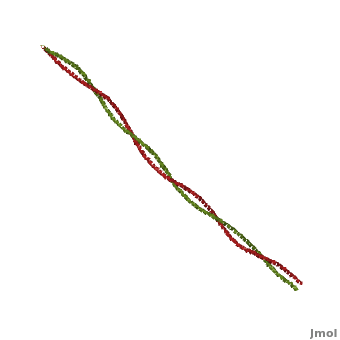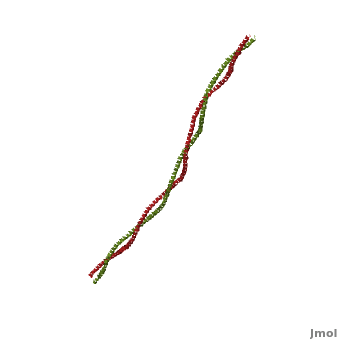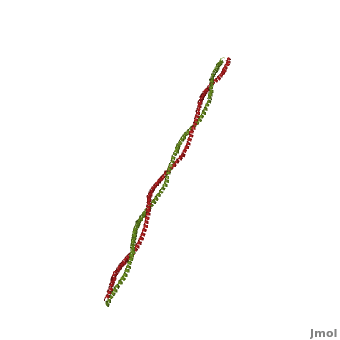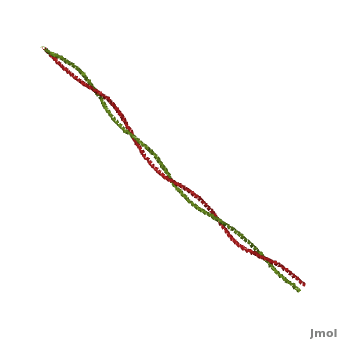
 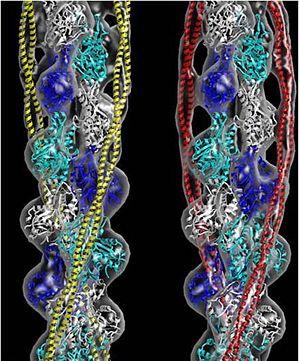 Tropomyosin (TM) is an actin binding protein, which consists of a coiled-coil dimer (see left) and forms a polymer along the length of actin by a head-to-tail overlap along the major grove of actin (see down & left)[1]. The head-to-tail overlap allows flexibility between the tropomyosin dimers so it will lay unstrained along the filament[1]. Each tropomyosin molecule spans seven actin monomers within a filament and lays N- to C- terminally from actin's pointed to barbed end[2]. The 284 amino acid helix has a length of 420 Angstroms and has a molecular weight around 65-70 kilodaltons (vertebrate tropomyosin)[1][3]. A few of tropomyosin's characteristics as an actin binding protein includes regulation, stabilization and recruitment.
Tropomyosin's Structure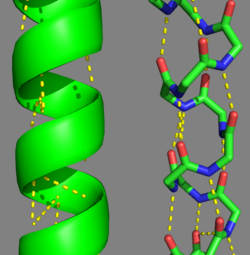 As determined by the Structural Classification of Protein's (SCOP) Database, Tropomyosin is categorized as follows (general to specific):
 Categorization of tropomyosin's describes a unique pattern of amino acids within the primary structure of the alpha helices that comprise the dimer interface[1]. The unique amino acid pattern, found within all coiled-coil proteins, is a heptad repeat, which follows a similar pattern to: H-P-P-H-C-P-C, where H is hydrophobic, P is polar and C is charged[1][3]. This heptad repeat forms a right-handed alpha helical secondary structure (see right for alpha helix secondary structure)[1]. This alpha helix is special in that it forms a hydrophobic strip along one side, which will interface with an adjacent alpha helix that also contains the heptad repeat and hydrophobic strip. These strips aid in the dimerization of tropomyosin and is important in the characteristic coiled-coil domain.
Post-Translational ModificationsThere are two types of post-translational modifications to tropomyosin: phosphorylation acetylation[1]. Phosphorylation occurs on amino acid [1]. This phosphorylation is occurs as a result of oxidative stress, which is associated with actin remodeling and recruitment of additional tropomyosin into stress fibers[1]. The acetylation occurs on the N-terminus of the N-terminal methionine, which is essential for: coiled-coil stability, overlap formation and actin binding[2][1]. Evolutionary ConservationTropomyosin is highly conserved actin binding protein, which is found in Eukarya from the animal kingdom to yeast, with the exception of plants[1]. The earliest characterization of tropomyosin gene lineage was in yeast (budding and fission yeast)[1][6]. These genes are known as TPM1 and TPM2, respectively, and share 64.5% sequence identity[1][6]. As we move away from unicellular organisms and into multicellular invertebrates, tropomyosin genes in nematodes slightly diverge from yeast, but have 85-90% sequence identity between their genes[1]. Further analysis for vertebrates show there are four genes that generate over 40 known mammalian isoforms of tropomyosin, which are synthesized by exon splicing[1]. The slight evolution change of tropomyosin has occurred as a result of the increasing need of tropomyosin to function in different systems, but tropomyosin has evolutionarily stayed well conserved because of the basic structural pressures imposed on the protein[1]. It is interesting to note, the region with the least conservation has been in the N and C terminus. This is the result of head-tail interactions changing to accommodate different polymer confirmations along actin for different functions[1]. (To see this evolutionary conservation, go to the top right image of the web page and click on the "show" link, which is to the right of "Evolutionary Conservation".)
Tropomyosin's Function: Muscle and Non-Muscle SystemsMuscle tissue is comprised of many muscle fibers or cells. Those muscle fibers consist of myofibrils that contain a series of contractile units called sarcomeres. Within this unit contains thick filaments, comprised mainly of myosin, and thin filaments, which contain actin, tropomyosin and troponin. Tropomyosin in striated muscle systems (skeletal and cardiac) acts to inhibit the myosin cross-bridges from binding to the myosin binding site on thin filaments, this tropomyosin state is in the "Blocked" position[7]. When the muscle is stimulated, there is a rise in intracellular calcium stemming from a cascade of cellular processes. As the calcium is bathing the sarcomere, it will bind to the troponin complex, which is bound to both actin and tropomyosin[8]. The troponin will displace the tropomyosin from a "Blocked" to a "Closed" position[7]. This transition allows the myosin head to interact weakly with the myosin binding site[8]. The tropomyosin is displaced to its final position, "Open" state, along actin filament as myosin binds to its site[7]. These three tropomyosin states along the filament is referred to as the three state model[7]. As the intracellular calcium concentration falls, the troponin no longer is able to displace tropomyosin and it will transition back to the "Blocked" state[7].
|
| ||||||||||
3D structures of Tropomyosin3D structures of Tropomyosin
Updated on 25-May-2014
3mtu, 3mud – cTPM alpha-1 – chicken
1ic2 - cTPM alpha-1 (mutant)
3u1a, 3u1c – cTPM α-1 N terminal
3u59 – cTPM β N terminal
2w49, 2w4u – cTnnC+cTnnT+cTnnI+cTPM alpha-1+cActin
2z5h – yTPM alpha-1 N-terminal+C-terminal+GNC4 leucine zipper+TnnT – yeast
2z5i - yTPM alpha-1 N-terminal+C-terminal+GNC4 leucine zipper
2efr, 2efs, 2d3e - raTPM alpha-1 C-terminal+GNC4 leucine zipper – rabbit
4a7f, 4a7h, 4a7l - raTPM α-1 + myosin + actin – Cryo EM
3j4k - raTPM + actin – Cryo EM
1kql - TPM alpha-1 C-terminal+GNC4 leucine zipper - rat
1mv4 - rTPM alpha-1 C-terminal
2g9j - rTPM alpha-1 TM9A+GNC4
2b9c – rTPM mid region
3azd – rTPM N terminal
1c1g – TPM – pig
2tma – TPM - model
ReferencesReferences
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 1.21 1.22 1.23 1.24 Tropomyosins. I. Gunning, Peter, 1950- II. Series.[DNLM: 1. Tropomyosin. W1 AD559 v.644 2008 / WE 500 T856 2008]
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Frye J, Klenchin VA, Rayment I. Structure of the tropomyosin overlap complex from chicken smooth muscle: insight into the diversity of N-terminal recognition . Biochemistry. 2010 Jun 15;49(23):4908-20. PMID:20465283 doi:10.1021/bi100349a
- ↑ 3.0 3.1 3.2 Whitby FG, Phillips GN Jr. Crystal structure of tropomyosin at 7 Angstroms resolution. Proteins. 2000 Jan 1;38(1):49-59. PMID:10651038
- ↑ 4.0 4.1 4.2 Clayton JE, Sammons MR, Stark BC, Hodges AR, Lord M. Differential regulation of unconventional fission yeast myosins via the actin track. Curr Biol. 2010 Aug 24;20(16):1423-31. Epub 2010 Aug 12. PMID:20705471 doi:10.1016/j.cub.2010.07.026
- ↑ 5.0 5.1 Stark BC, Sladewski TE, Pollard LW, Lord M. Tropomyosin and myosin-II cellular levels promote actomyosin ring assembly in fission yeast. Mol Biol Cell. 2010 Mar 15;21(6):989-1000. Epub 2010 Jan 28. PMID:20110347 doi:10.1091/mbc.E09-10-0852
- ↑ 6.0 6.1 Drees B, Brown C, Barrell BG, Bretscher A. Tropomyosin is essential in yeast, yet the TPM1 and TPM2 products perform distinct functions. J Cell Biol. 1995 Feb;128(3):383-92. PMID:7844152
- ↑ 7.0 7.1 7.2 7.3 7.4 Lehman W, Galinska-Rakoczy A, Hatch V, Tobacman LS, Craig R. Structural basis for the activation of muscle contraction by troponin and tropomyosin. J Mol Biol. 2009 May 15;388(4):673-81. Epub 2009 Mar 31. PMID:19341744 doi:10.1016/j.jmb.2009.03.060
- ↑ 8.0 8.1 Tyska MJ, Warshaw DM. The myosin power stroke. Cell Motil Cytoskeleton. 2002 Jan;51(1):1-15. PMID:11810692 doi:10.1002/cm.10014