3fpl: Difference between revisions
No edit summary |
No edit summary |
||
| Line 2: | Line 2: | ||
===Chimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-295 of C. beijerinckii ADH by T. brockii ADH=== | ===Chimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-295 of C. beijerinckii ADH by T. brockii ADH=== | ||
{{ABSTRACT_PUBMED_20102159}} | {{ABSTRACT_PUBMED_20102159}} | ||
==Function== | |||
[[http://www.uniprot.org/uniprot/ADH_CLOBE ADH_CLOBE]] Alcohol dehydrogenase with a preference for medium chain secondary alcohols, such as 2-butanol and isopropanol. Has very low activity with primary alcohols, such as ethanol. Under physiological conditions, the enzyme reduces aldehydes and 2-ketones to produce secondary alcohols. Is active with acetaldehyde and propionaldehyde.<ref>PMID:8349550</ref> <ref>PMID:20102159</ref> | |||
==About this Structure== | ==About this Structure== | ||
[[3fpl]] is a 1 chain structure with sequence from [http://en.wikipedia.org/wiki/ | [[3fpl]] is a 1 chain structure with sequence from [http://en.wikipedia.org/wiki/"clostridium_rubrum"_ng_and_vaughn_1963 "clostridium rubrum" ng and vaughn 1963]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3FPL OCA]. | ||
==See Also== | ==See Also== | ||
| Line 14: | Line 17: | ||
==Reference== | ==Reference== | ||
<ref group="xtra">PMID:020102159</ref><references group="xtra"/> | <ref group="xtra">PMID:020102159</ref><references group="xtra"/><references/> | ||
[[Category: | [[Category: Clostridium rubrum ng and vaughn 1963]] | ||
[[Category: Burstein, Y.]] | [[Category: Burstein, Y.]] | ||
[[Category: Felix, F.]] | [[Category: Felix, F.]] | ||
Revision as of 08:51, 29 January 2014
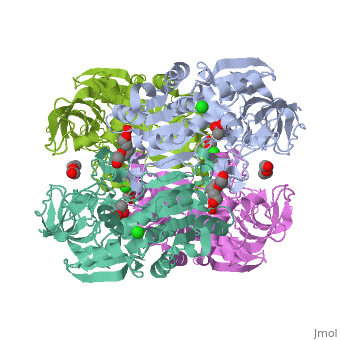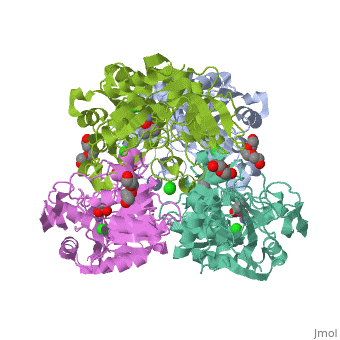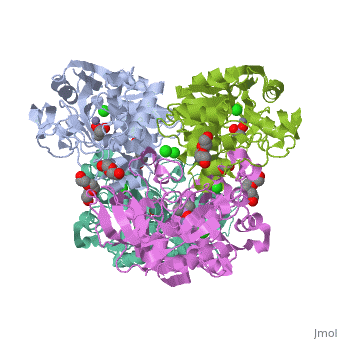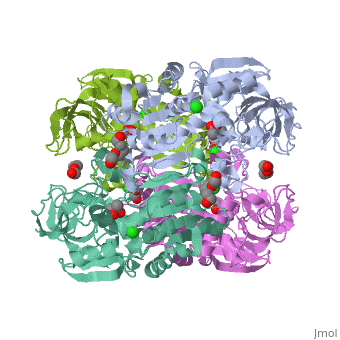
| |||||||
| 3fpl, resolution 1.90Å () | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , , , , | ||||||
| Gene: | ADH1, ADH1 ("Clostridium rubrum" Ng and Vaughn 1963) | ||||||
| Activity: | Alcohol dehydrogenase (NADP(+)), with EC number 1.1.1.2 | ||||||
| Related: | 3fpc | ||||||
| |||||||
| |||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
Chimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-295 of C. beijerinckii ADH by T. brockii ADHChimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-295 of C. beijerinckii ADH by T. brockii ADH
The cofactor-binding domains (residues 153-295) of the alcohol dehydrogenases from the thermophile Thermoanaerobacter brockii (TbADH), the mesophilic bacterium Clostridium beijerinckii (CbADH), and the protozoan parasite Entamoeba histolytica (EhADH1) have been exchanged. Three chimeras have been constructed. In the first chimera, the cofactor-binding domain of thermophilic TbADH was replaced with the cofactor-binding domain of its mesophilic counterpart CbADH [chimera Chi21((TCT))]. This domain exchange significantly destabilized the parent thermophilic enzyme (DeltaT(1/2) = -18 degrees C). The reverse exchange in CbADH [chimera Chi22((CTC))], however, had little effect on the thermal stability of the parent mesophilic protein. Furthermore, substituting the cofactor-binding domain of TbADH with the homologous domain of EhADH1 [chimera Chi23((TET))] substantially reduced the thermal stability of the thermophilic ADH (DeltaT(1/2) = -51 degrees C) and impeded the oligomerization of the enzyme. All three chimeric proteins and one of their site-directed mutants were crystallized, and their three-dimensional (3D) structures were determined. Comparison of the 3D structures of the chimeras and the chimeric mutant with the structures of their parent ADHs showed no significant changes to their Calpha chains, suggesting that the difference in the thermal stability of the three parent ADHs and their chimeric mutants could be due to a limited number of substitutions located at strategic positions, mainly at the oligomerization interfaces. Indeed, stabilization of the chimeras was achieved, to a significant extent, either by introduction of a proline residue at a strategic position in the major horse liver ADH-type dimerization interface (DeltaT(1/2) = 35 degrees C) or by introduction of intersubunit electrostatic interactions (DeltaT(1/2) = 6 degrees C).
Biochemical and Structural Properties of Chimeras Constructed by Exchange of Cofactor-Binding Domains in Alcohol Dehydrogenases from Thermophilic and Mesophilic Microorganisms., Goihberg E, Peretz M, Tel-Or S, Dym O, Shimon L, Frolow F, Burstein Y, Biochemistry. 2010 Feb 9. PMID:20102159
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
FunctionFunction
[ADH_CLOBE] Alcohol dehydrogenase with a preference for medium chain secondary alcohols, such as 2-butanol and isopropanol. Has very low activity with primary alcohols, such as ethanol. Under physiological conditions, the enzyme reduces aldehydes and 2-ketones to produce secondary alcohols. Is active with acetaldehyde and propionaldehyde.[1] [2]
About this StructureAbout this Structure
3fpl is a 1 chain structure with sequence from "clostridium_rubrum"_ng_and_vaughn_1963 "clostridium rubrum" ng and vaughn 1963. Full crystallographic information is available from OCA.
See AlsoSee Also
- Alcohol dehydrogenase
- Chimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-295 of C. beijerinckii ADH by T. brockii ADH
- Chimeras of alcohol dehydrogenases
- Q165E/S254K Double Mutant Chimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-295 of T. brockii ADH by C. beijerinckii ADH
- Tetrameric alcohol dehydrogenases
ReferenceReference
- ↑ Goihberg E, Peretz M, Tel-Or S, Dym O, Shimon L, Frolow F, Burstein Y. Biochemical and Structural Properties of Chimeras Constructed by Exchange of Cofactor-Binding Domains in Alcohol Dehydrogenases from Thermophilic and Mesophilic Microorganisms. Biochemistry. 2010 Feb 9. PMID:20102159 doi:10.1021/bi901730x
- ↑ Ismaiel AA, Zhu CX, Colby GD, Chen JS. Purification and characterization of a primary-secondary alcohol dehydrogenase from two strains of Clostridium beijerinckii. J Bacteriol. 1993 Aug;175(16):5097-105. PMID:8349550
- ↑ Goihberg E, Peretz M, Tel-Or S, Dym O, Shimon L, Frolow F, Burstein Y. Biochemical and Structural Properties of Chimeras Constructed by Exchange of Cofactor-Binding Domains in Alcohol Dehydrogenases from Thermophilic and Mesophilic Microorganisms. Biochemistry. 2010 Feb 9. PMID:20102159 doi:10.1021/bi901730x