Molecular Playground/CsoR and RcnR: Difference between revisions
Carolyn Carr (talk | contribs) No edit summary |
Carolyn Carr (talk | contribs) No edit summary |
||
| Line 24: | Line 24: | ||
The Ni(II) site is bound by the N-terminal amine, Cys35, and His64.<ref>PMID: 23215580</ref><ref>PMID: 22471551</ref> Because the Ni(II) site has an octahedral geometry, there are three metal binding residues whose identity is unknown. However, using XAS and lacZ activity assays it is known that the remaining ligands bind to the Ni(II) using N/O atoms and that there are a total of two histidine ligand bound to the Ni(II), leaving the identity of one unaccounted for. Studies have shown that this histidine is not His3, His60, or His67. | The Ni(II) site is bound by the N-terminal amine, Cys35, and His64.<ref>PMID: 23215580</ref><ref>PMID: 22471551</ref> Because the Ni(II) site has an octahedral geometry, there are three metal binding residues whose identity is unknown. However, using XAS and lacZ activity assays it is known that the remaining ligands bind to the Ni(II) using N/O atoms and that there are a total of two histidine ligand bound to the Ni(II), leaving the identity of one unaccounted for. Studies have shown that this histidine is not His3, His60, or His67. | ||
[[Image:Ni Site.png|200px|thumb|Fig. 1: Schematic of known Ni(II) binding residues.]] | |||
The Co(II) site is bound by the the N-terminal amine, His3, Cys35, His64 and possibly His60. The Co(II) site of RcnR also has an octahedral geometry, leaving 1-2 residues whose identity is unknown. XAS reveals that these ligands are bound to Co(II) using N/O atoms and that there are a total of three histidine ligands bound to the Co(II). Studies have shown that His67 is not a Co(II) ligand. | The Co(II) site is bound by the the N-terminal amine, His3, Cys35, His64 and possibly His60. The Co(II) site of RcnR also has an octahedral geometry, leaving 1-2 residues whose identity is unknown. XAS reveals that these ligands are bound to Co(II) using N/O atoms and that there are a total of three histidine ligands bound to the Co(II). Studies have shown that His67 is not a Co(II) ligand. | ||
== Research Interests == | == Research Interests == | ||
Revision as of 20:12, 30 October 2013
|
One of the CBI Molecules being studied in the University of Massachusetts Amherst Chemistry-Biology Interface Program at UMass Amherst and on display at the Molecular Playground.
IntroductionIntroduction
Heavy metals such as iron, nickel, copper, and zinc are important cofactors for the functions of many different metalloenzymes. High levels of these heavy metals can also cause damage to cellular components, therefore intracellular levels of metals are tightly regulated within the cell. One of the ways that bacteria can regulate intracellular metal levels is by increasing the amount of metal efflux proteins. CsoR and RcnR are members of a large family of metal-responsive DNA-binding proteins, both of which regulate the transcription of metal-specific efflux proteins. CsoR is only responsive to the binding of Cu(I); whereas RcnR is only responsive to the binding of Ni(II) or Co(II).[1]
RcnR and CsoRRcnR and CsoR
In Escherichia coli apo-RcnR blocks the transcription of nickel and cobalt efflux proteins RcnA and RcnB by binding to its promoter region. Although no crystal structure of RcnR is available on the PDB, the Cu(I)-bound CsoR crystal structure from Mycobacterium tuberculosis is available and RcnR is predicted to share a similar fold to CsoR. Upon Ni(II)- or Co(II)-binding, RcnR is released from DNA allowing the transcription of RcnA and RcnB, facilitating the efflux of Ni(II) and Co(II).[2] CsoR has been characterized in Bacillus subtilis[3] and M. tuberculosis[4] to release from the promoter regions of copper-efflux operons upon binding of Cu(I). The analogous functions of CsoR and RcnR in addition to local sequence similarity (25% identical, 56% similar) suggests a conserved mode of function in this family of metal-responsive DNA-binding proteins.[5]
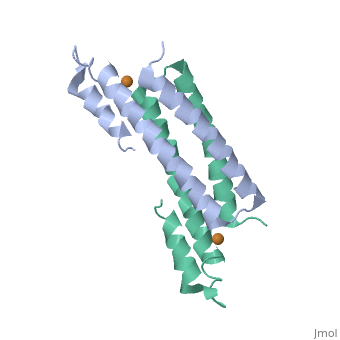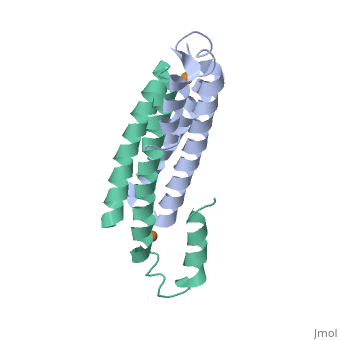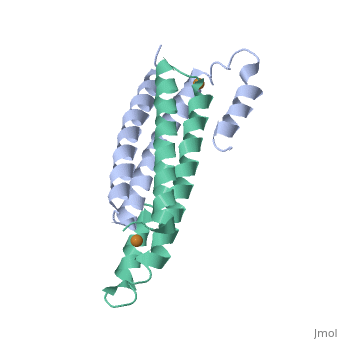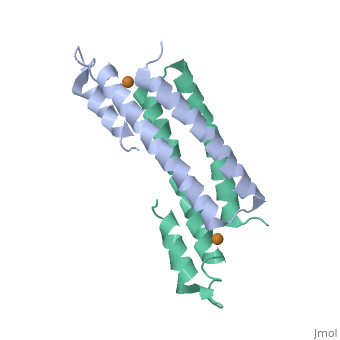
Structure of CsoR and RcnRStructure of CsoR and RcnR
binds one Cu(I) per monomer. The protein forms a dimer of dimers with a in the tetrameric interface. Each of a dimer unit. Where one monomer is bound to Cu(I) by His61 and Cys65, the other monomer is bound to the metal by Cys35.[6] E. coli
RcnR is also tetrameric and has the same protein-to-metal stoichiometry. Through a combination of site-directed mutagenesis, lacZ activity assays and X-ray absorption spectroscopy (XAS) structural studies it is now known that RcnR uses a different ligand set to bind Ni(II) than it does Co(II).
The Ni(II) site is bound by the N-terminal amine, Cys35, and His64.[7][8] Because the Ni(II) site has an octahedral geometry, there are three metal binding residues whose identity is unknown. However, using XAS and lacZ activity assays it is known that the remaining ligands bind to the Ni(II) using N/O atoms and that there are a total of two histidine ligand bound to the Ni(II), leaving the identity of one unaccounted for. Studies have shown that this histidine is not His3, His60, or His67.
The Co(II) site is bound by the the N-terminal amine, His3, Cys35, His64 and possibly His60. The Co(II) site of RcnR also has an octahedral geometry, leaving 1-2 residues whose identity is unknown. XAS reveals that these ligands are bound to Co(II) using N/O atoms and that there are a total of three histidine ligands bound to the Co(II). Studies have shown that His67 is not a Co(II) ligand.
Research InterestsResearch Interests
The mechanism of DNA binding of the CsoR/RcnR family of metal-responsive transcriptional regulators is still unknown. Additionally, RcnR has an added level of complexity because it is reponsive to both Ni(II) and Co(II) binding. The Maroney Lab at the University of Massachusetts Amherst is interested in the conformational changes of RcnR induced by DNA-, Ni(II)-, and Co(II)-binding. Identification of the remaining metal binding residues in RcnR is ongoing, in addition to identification of the DNA-binding residues in RcnR, as there is no crystal structure of any member of this family of proteins binding to DNA.
3D structures of copper homeostasis protein3D structures of copper homeostasis protein
ReferencesReferences
- ↑ Reyes-Caballero H, Campanello GC, Giedroc DP. Metalloregulatory proteins: metal selectivity and allosteric switching. Biophys Chem. 2011 Jul;156(2-3):103-14. Epub 2011 Apr 5. PMID:21511390 doi:10.1016/j.bpc.2011.03.010
- ↑ Iwig JS, Chivers PT. Coordinating intracellular nickel-metal-site structure-function relationships and the NikR and RcnR repressors. Nat Prod Rep. 2010 May;27(5):658-67. Epub 2010 Mar 5. PMID:20442957 doi:10.1039/b906683g
- ↑ Smaldone GT, Helmann JD. CsoR regulates the copper efflux operon copZA in Bacillus subtilis. Microbiology. 2007 Dec;153(Pt 12):4123-8. PMID:18048925 doi:10.1099/mic.0.2007/011742-0
- ↑ Liu T, Ramesh A, Ma Z, Ward SK, Zhang L, George GN, Talaat AM, Sacchettini JC, Giedroc DP. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat Chem Biol. 2007 Jan;3(1):60-8. Epub 2006 Dec 3. PMID:17143269 doi:http://dx.doi.org/10.1038/nchembio844
- ↑ Iwig JS, Leitch S, Herbst RW, Maroney MJ, Chivers PT. Ni(II) and Co(II) sensing by Escherichia coli RcnR. J Am Chem Soc. 2008 Jun 18;130(24):7592-606. Epub 2008 May 28. PMID:18505253 doi:10.1021/ja710067d
- ↑ Liu T, Ramesh A, Ma Z, Ward SK, Zhang L, George GN, Talaat AM, Sacchettini JC, Giedroc DP. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat Chem Biol. 2007 Jan;3(1):60-8. Epub 2006 Dec 3. PMID:17143269 doi:http://dx.doi.org/10.1038/nchembio844
- ↑ Higgins KA, Hu HQ, Chivers PT, Maroney MJ. Effects of select histidine to cysteine mutations on transcriptional regulation by Escherichia coli RcnR. Biochemistry. 2013 Jan 8;52(1):84-97. doi: 10.1021/bi300886q. Epub 2012 Dec 24. PMID:23215580 doi:http://dx.doi.org/10.1021/bi300886q
- ↑ Higgins KA, Chivers PT, Maroney MJ. Role of the N-terminus in determining metal-specific responses in the E. coli Ni- and Co-responsive metalloregulator, RcnR. J Am Chem Soc. 2012 Apr 25;134(16):7081-93. doi: 10.1021/ja300834b. Epub 2012 Apr, 11. PMID:22471551 doi:http://dx.doi.org/10.1021/ja300834b