Single stranded binding protein: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
== Sandbox Single Stranded DNA-Binding Protein (SSB)== | == Sandbox Single Stranded DNA-Binding Protein (SSB)== | ||
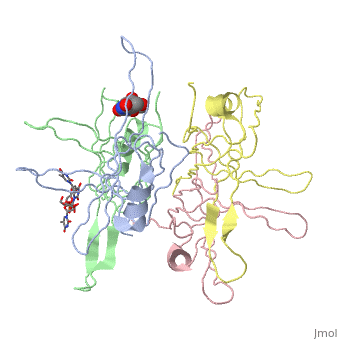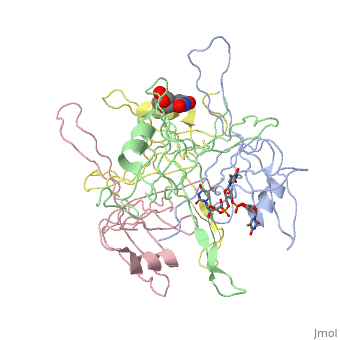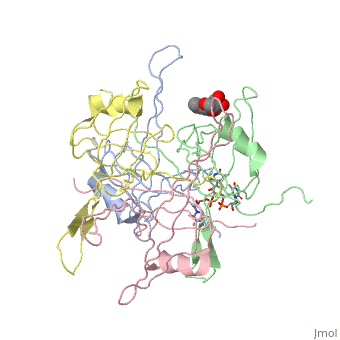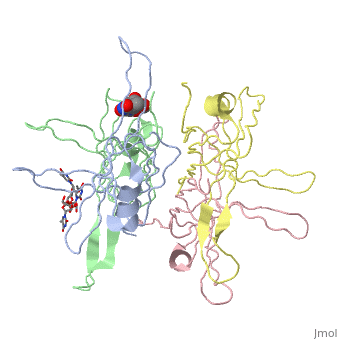
<StructureSection load='1eqq' size=' | <StructureSection load='1eqq' size='540' side='right' frame='true' align='right' caption='Structure of Single Stranded DNA-Binding Protein (PDB entry [[1eqq]])' scene=''> | ||
<StructureSection load='1kaw' size='350' side='right' caption='Structure of Single Stranded DNA-Binding Protein (PDB entry [[1kaw]])' scene=''> | <StructureSection load='1kaw' size='350' side='right' caption='Structure of Single Stranded DNA-Binding Protein (PDB entry [[1kaw]])' scene=''> | ||
Revision as of 17:50, 30 October 2013
Sandbox Single Stranded DNA-Binding Protein (SSB)Sandbox Single Stranded DNA-Binding Protein (SSB)
<StructureSection load='1kaw' size='350' side='right' caption='Structure of Single Stranded DNA-Binding Protein (PDB entry 1kaw)' scene=> IntroductionThe single-stranded DNA-binding protein (SSB) of Escherichia coli is involved in all aspects of DNA metabolism: replication, repair, and recombination. In solution, the protein exists as a homotetramer of 18,843-kilodalton subunits. As it binds tightly and cooperatively to single-stranded DNA, it has become a prototypic model protein for studying protein-nucleic acid interactions. The sequences of the gene and protein are known, and the functional domains of subunit interaction, DNA binding, and protein-protein interactions have been probed by structure-function analyses of various mutations. The ssb gene has three promoters, one of which is inducible because it lies only two nucleotides from the LexA-binding site of the adjacent uvrA gene. Induction of the SOS response, however, does not lead to significant increases in SSB levels. The binding protein has several functions in DNA replication, including enhancement of helix destabilization by DNA helicases, prevention of reannealing of the single strands and protection from nuclease digestion, organization and stabilization of replication origins, primosome assembly, priming specificity, enhancement of replication fidelity, enhancement of polymerase processivity, and promotion of polymerase binding to the template. E. coli SSB is required for methyl-directed mismatch repair, induction of the SOS response, and recombinational repair. During recombination, SSB interacts with the RecBCD enzyme to find Chi sites, promotes binding of RecA protein, and promotes strand uptake.
References
PMID: 2087220
|
| ||||||||||