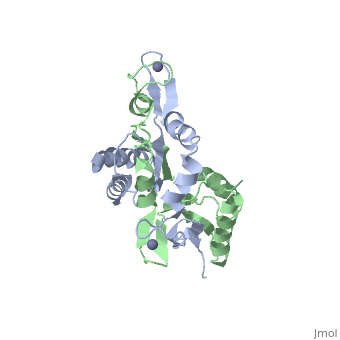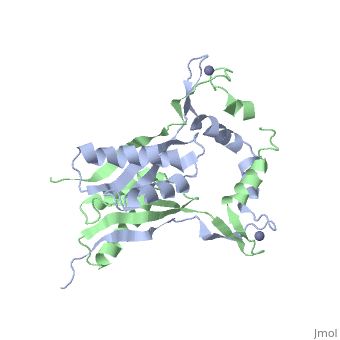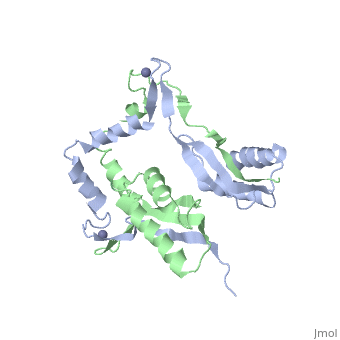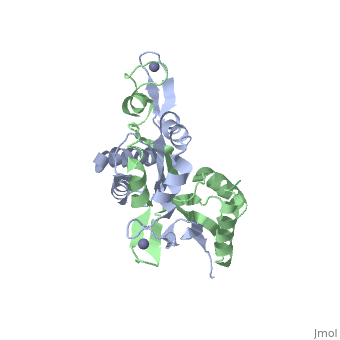Heidi Hu/Sandbox 2: Difference between revisions
No edit summary |
No edit summary |
||
| Line 11: | Line 11: | ||
[http://en.wikipedia.org/wiki/Helicobacter_pylori ''Helicobacter pylori''] is a pathogenic bacterium that colonizes the human gastric mucosa, which can cause peptic ulcers and has been linked to stomach cancers.<ref>PMID:9146793</ref> [http://en.wikipedia.org/wiki/Helicobacter_pylori ''H. pylori''] requires the activity of nickel-dependent enzymes, [NiFe]-[http://en.wikipedia.org/wiki/Hydrogenase ''Hydrogenase''] (H<sub>2</sub>ase) and [http://en.wikipedia.org/wiki/Urease ''urease''], to survive in the acidic environment of the stomach.<ref>PMID:2050411</ref><ref>PMID:8063376</ref><ref>PMID:12459589</ref> Thus nickel is an important nutrient for H. pylori. | [http://en.wikipedia.org/wiki/Helicobacter_pylori ''Helicobacter pylori''] is a pathogenic bacterium that colonizes the human gastric mucosa, which can cause peptic ulcers and has been linked to stomach cancers.<ref>PMID:9146793</ref> [http://en.wikipedia.org/wiki/Helicobacter_pylori ''H. pylori''] requires the activity of nickel-dependent enzymes, [NiFe]-[http://en.wikipedia.org/wiki/Hydrogenase ''Hydrogenase''] (H<sub>2</sub>ase) and [http://en.wikipedia.org/wiki/Urease ''urease''], to survive in the acidic environment of the stomach.<ref>PMID:2050411</ref><ref>PMID:8063376</ref><ref>PMID:12459589</ref> Thus nickel is an important nutrient for H. pylori. | ||
Accessory proteins HypABCDEF and UreIEFGH facilitate in the maturation of [NiFe]-[http://en.wikipedia.org/wiki/Hydrogenase ''H<sub>2</sub>ase''] and [http://en.wikipedia.org/wiki/Urease ''urease''] respectively. HypA is a nickel metallochaperone normally associated with the maturation of [NiFe]-[http://en.wikipedia.org/wiki/Hydrogenase ''H<sub>2</sub>ase'']. In H. pylori, however, it is also require for the full activy of urease, despite the presence of the urease-specific Ni-chaperone, UreE. | Accessory proteins HypABCDEF and UreIEFGH facilitate in the maturation of [NiFe]-[http://en.wikipedia.org/wiki/Hydrogenase ''H<sub>2</sub>ase''] and [http://en.wikipedia.org/wiki/Urease ''urease''] respectively<ref>PMID:</ref>. HypA is a nickel metallochaperone normally associated with the maturation of [NiFe]-[http://en.wikipedia.org/wiki/Hydrogenase ''H<sub>2</sub>ase'']<ref>PMID:</ref>. In H. pylori, however, it is also require for the full activy of urease, despite the presence of the urease-specific Ni-chaperone, UreE<ref>PMID:</ref>. | ||
| Line 21: | Line 21: | ||
caption='NMR structure of monomeric H. pylori HypA (PDB ID: [http://www.rcsb.org/pdb/explore/explore.do?structureId=3A44 3A44])' /> | caption='NMR structure of monomeric H. pylori HypA (PDB ID: [http://www.rcsb.org/pdb/explore/explore.do?structureId=3A44 3A44])' /> | ||
<br> | <br> | ||
The [http://en.wikipedia.org/wiki/Helicobacter_pylori ''H. pylori'']HypA (''Hp''HypA) is a 13.2kDa Ni-chaperone with both a nickel binding site and and structural zinc site. Zn(II) is coordinated by two | The [http://en.wikipedia.org/wiki/Helicobacter_pylori ''H. pylori'']HypA (''Hp''HypA) is a 13.2kDa Ni-chaperone with both a nickel binding site and and structural zinc site. Zn(II) is coordinated by two CXXC motif each with a flanking histidine, whereas the Ni(II) is known to bind to the N-terminus MHE motif<ref>PMID:</ref>. The ''Hp''HypA protein has also been characterized as a monomer or a homodimer. The N-terminal modified monomeric structure has been solved by NMR (PDB ID: [http://www.rcsb.org/pdb/explore/explore.do?structureId=2KDX 2KDX])<ref>PMID:</ref>. The monomeric structure shows an alpha/beta lobe containing the N- and C-termini well separated from the zinc binding lobe (Fig. 1). The homodimeric ''Hp''HypA has been characterized by NMR to have the similar overall structure with exception to the metal binding sites<ref>PMID:</ref>. The metal sites in ''Hp''HypA have been characterized by [http://en.wikipedia.org/wiki/X-ray_absorption_spectroscopy ''X-ray absorption spectroscopy''] (XAS) at pH 6.3 and 7.2 similating the internal pH of H. pylori under acid shock or at neutral pH conditions. Whereas the Ni(II) site is 6-coordinate N/O under both pH conditions, the Zn(II) coordination changes from Cys<sub>4</sub> at neutral pH to Cys<sub>2</sub>His<sub>2</sub> at acidic pH with nickel-bound<ref>PMID:</ref>. Changes in the coordination of structural Zinc site in response to pH has been hypothesized to be linked to altered HypA structures and protein interaction partners<ref>PMID:</ref>. | ||
The crystal structure of monomeric (PDB ID: [http://www.rcsb.org/pdb/explore/explore.do?structureId=3A43 3A43]) and homodimeric (PDB ID: [http://www.rcsb.org/pdb/explore/explore.do?structureId=3A44 3A44]) HypA from [http://en.wikipedia.org/wiki/Thermococcus_kodakarensis ''Thermococcus kodakarensis''] has been solved. The HypA homodimer from [http://en.wikipedia.org/wiki/Thermococcus_kodakarensis ''T. kodakarensis''] shows a switch dimer, where one beta strand of each alpha/beta lobe comes from the opposite subunit. Additionally, each Zn(II) site is coordinated by CXXC motifs from a different subunit. These observations were consistent with the [http://en.wikipedia.org/wiki/Helicobacter_pylori ''H. pylori''] homodimeric HypA NMR data. | The crystal structure of monomeric (PDB ID: [http://www.rcsb.org/pdb/explore/explore.do?structureId=3A43 3A43]) and homodimeric (PDB ID: [http://www.rcsb.org/pdb/explore/explore.do?structureId=3A44 3A44]) HypA from [http://en.wikipedia.org/wiki/Thermococcus_kodakarensis ''Thermococcus kodakarensis''] has been solved<ref>PMID:</ref>. The HypA homodimer from [http://en.wikipedia.org/wiki/Thermococcus_kodakarensis ''T. kodakarensis''] shows a switch dimer, where one beta strand of each alpha/beta lobe comes from the opposite subunit. Additionally, each Zn(II) site is coordinated by CXXC motifs from a different subunit. These observations were consistent with the [http://en.wikipedia.org/wiki/Helicobacter_pylori ''H. pylori''] homodimeric HypA NMR data. | ||
Revision as of 02:04, 13 December 2012
One of the CBI Molecules being studied in the University of Massachusetts Amherst Chemistry-Biology Interface Program at UMass Amherst and on display at the Molecular Playground.
IntroductionIntroduction
Helicobacter pylori is a pathogenic bacterium that colonizes the human gastric mucosa, which can cause peptic ulcers and has been linked to stomach cancers.[1] H. pylori requires the activity of nickel-dependent enzymes, [NiFe]-Hydrogenase (H2ase) and urease, to survive in the acidic environment of the stomach.[2][3][4] Thus nickel is an important nutrient for H. pylori.
Accessory proteins HypABCDEF and UreIEFGH facilitate in the maturation of [NiFe]-H2ase and urease respectively[5]. HypA is a nickel metallochaperone normally associated with the maturation of [NiFe]-H2ase[6]. In H. pylori, however, it is also require for the full activy of urease, despite the presence of the urease-specific Ni-chaperone, UreE[7].
HypAHypA
|
The H. pyloriHypA (HpHypA) is a 13.2kDa Ni-chaperone with both a nickel binding site and and structural zinc site. Zn(II) is coordinated by two CXXC motif each with a flanking histidine, whereas the Ni(II) is known to bind to the N-terminus MHE motif[8]. The HpHypA protein has also been characterized as a monomer or a homodimer. The N-terminal modified monomeric structure has been solved by NMR (PDB ID: 2KDX)[9]. The monomeric structure shows an alpha/beta lobe containing the N- and C-termini well separated from the zinc binding lobe (Fig. 1). The homodimeric HpHypA has been characterized by NMR to have the similar overall structure with exception to the metal binding sites[10]. The metal sites in HpHypA have been characterized by X-ray absorption spectroscopy (XAS) at pH 6.3 and 7.2 similating the internal pH of H. pylori under acid shock or at neutral pH conditions. Whereas the Ni(II) site is 6-coordinate N/O under both pH conditions, the Zn(II) coordination changes from Cys4 at neutral pH to Cys2His2 at acidic pH with nickel-bound[11]. Changes in the coordination of structural Zinc site in response to pH has been hypothesized to be linked to altered HypA structures and protein interaction partners[12].
The crystal structure of monomeric (PDB ID: 3A43) and homodimeric (PDB ID: 3A44) HypA from Thermococcus kodakarensis has been solved[13]. The HypA homodimer from T. kodakarensis shows a switch dimer, where one beta strand of each alpha/beta lobe comes from the opposite subunit. Additionally, each Zn(II) site is coordinated by CXXC motifs from a different subunit. These observations were consistent with the H. pylori homodimeric HypA NMR data.
Research InterestsResearch Interests

The Ni- and pH-dependent changes in the H. pylori HypA structural zinc site suggests multiple conformations of this protein. Thus HypA is likely interfacing between the maturation of [NiFe]-H2ase and/or urease in H. pylori in a pH-dependent manner, as it is required for the full activation of both of these nickel enzymes. The Maroney Lab at the University of Massachusetts Amherst is interested in characterizing the conformational changes in HypA and its interaction partners under neutral and acid shock conditions.
ReferencesReferences
- ↑ Kuipers EJ. Helicobacter pylori and the risk and management of associated diseases: gastritis, ulcer disease, atrophic gastritis and gastric cancer. Aliment Pharmacol Ther. 1997 Apr;11 Suppl 1:71-88. PMID:9146793
- ↑ Eaton KA, Brooks CL, Morgan DR, Krakowka S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun. 1991 Jul;59(7):2470-5. PMID:2050411
- ↑ Eaton KA, Krakowka S. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect Immun. 1994 Sep;62(9):3604-7. PMID:8063376
- ↑ Olson JW, Maier RJ. Molecular hydrogen as an energy source for Helicobacter pylori. Science. 2002 Nov 29;298(5599):1788-90. PMID:12459589 doi:10.1126/science.1077123
- ↑ Jmol - a paradigm shift in crystallographic visualization. J. Appl. Cryst. (2010). 43, 1250-1260 doi:https://dx.doi.org/10.1107/S0021889810030256
- ↑ Jmol - a paradigm shift in crystallographic visualization. J. Appl. Cryst. (2010). 43, 1250-1260 doi:https://dx.doi.org/10.1107/S0021889810030256
- ↑ Jmol - a paradigm shift in crystallographic visualization. J. Appl. Cryst. (2010). 43, 1250-1260 doi:https://dx.doi.org/10.1107/S0021889810030256
- ↑ Jmol - a paradigm shift in crystallographic visualization. J. Appl. Cryst. (2010). 43, 1250-1260 doi:https://dx.doi.org/10.1107/S0021889810030256
- ↑ Jmol - a paradigm shift in crystallographic visualization. J. Appl. Cryst. (2010). 43, 1250-1260 doi:https://dx.doi.org/10.1107/S0021889810030256
- ↑ Jmol - a paradigm shift in crystallographic visualization. J. Appl. Cryst. (2010). 43, 1250-1260 doi:https://dx.doi.org/10.1107/S0021889810030256
- ↑ Jmol - a paradigm shift in crystallographic visualization. J. Appl. Cryst. (2010). 43, 1250-1260 doi:https://dx.doi.org/10.1107/S0021889810030256
- ↑ Jmol - a paradigm shift in crystallographic visualization. J. Appl. Cryst. (2010). 43, 1250-1260 doi:https://dx.doi.org/10.1107/S0021889810030256
- ↑ Jmol - a paradigm shift in crystallographic visualization. J. Appl. Cryst. (2010). 43, 1250-1260 doi:https://dx.doi.org/10.1107/S0021889810030256