RA Mediated T-reg Differentiation: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
==Introduction== | ==Introduction== | ||
T-regulatory cells (T-regs) are a small subset of CD4+ T-cells that exhibit strong down regulation of immune system activity in their local environment. They are distinguished from other CD4+ T-cells by the expression of FOXP3, a gene regulator. <ref> PMID: 19410687 </ref> These cells have been shown to differentiate from CD4+ T-helper cells upon activation and exposure to the following cytokines: tumor growth factor β (TGF-β), Interleukin-2 (IL-2) and retinoic acid (RA). <ref> PMID: 21839265 </ref> Both TGF-β and IL-2 are used in other immune system differentiation, however, RA has been shown to bias T-cells to the T-reg phenotype. <ref> PMID: 21839265 </ref> When acting upon T-reg cells, RA acts as the ligand for the Retinoic Acid Receptor-α (RARα) / Retinoid X Receptor-α (RXRα) heterodimer. This heterodimer is of the nuclear receptor family, and each chain consists of the same three part structure: a Ligand binding domain (LBD), a DNA binding domain (DBD), and a hinge region connecting the two binding domains. <ref> PMID: 10406480 </ref> | T-regulatory cells (T-regs) are a small subset of CD4+ T-cells that exhibit strong down regulation of immune system activity in their local environment. They are distinguished from other CD4+ T-cells by the expression of FOXP3, a gene regulator. <ref> PMID: 19410687 </ref> The exact mechanisms used by T-regs to down regulate the immune system has not yet been clearly elucidated. These cells have been shown to differentiate from CD4+ T-helper cells upon activation and exposure to the following cytokines: tumor growth factor β (TGF-β), Interleukin-2 (IL-2) and retinoic acid (RA). <ref> PMID: 21839265 </ref> Both TGF-β and IL-2 are used in other immune system differentiation, however, RA has been shown to bias T-cells to the T-reg phenotype. <ref> PMID: 21839265 </ref> When acting upon T-reg cells, RA acts as the ligand for the Retinoic Acid Receptor-α (RARα) / Retinoid X Receptor-α (RXRα) heterodimer. This heterodimer is of the nuclear receptor family, and each chain consists of the same three part structure: a Ligand binding domain (LBD), a DNA binding domain (DBD), and a hinge region connecting the two binding domains. <ref> PMID: 10406480 </ref> | ||
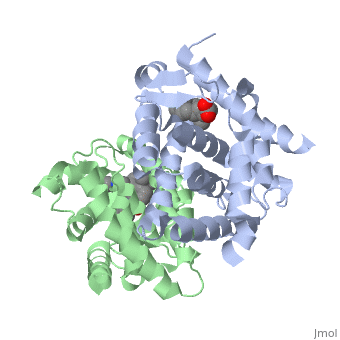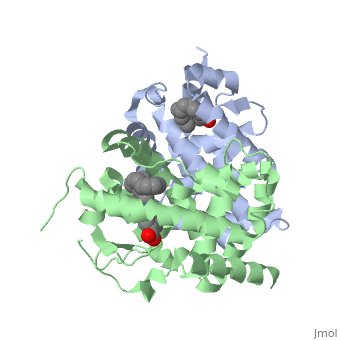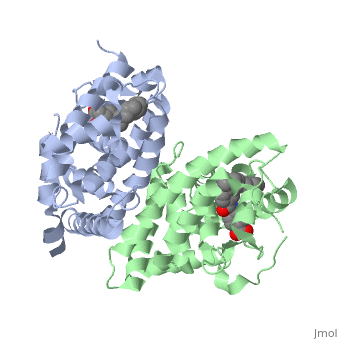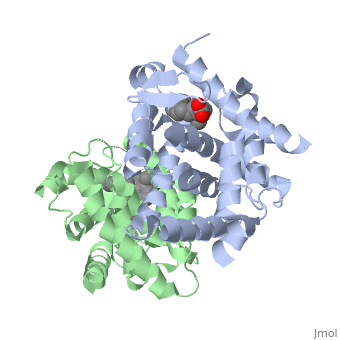
<StructureSection load='1dkf' size='350' side='left' caption='RARα-RXRα interaction (PDB entry [[1dkf]])' scene=''> | <StructureSection load='1dkf' size='350' side='left' caption='RARα-RXRα interaction (PDB entry [[1dkf]])' scene=''> | ||
==Ligand Binding Domain== | ==Ligand Binding Domain== | ||
| Line 10: | Line 10: | ||
The residues of <scene name='RA_Mediated_T-reg_Differentiaition/Rar_dimer_interface/ | The residues of <scene name='RA_Mediated_T-reg_Differentiaition/Rar_dimer_interface/2'> RAR-</scene>α that are interacting in the heterodimer are as follows: | ||
Hydrophobic residues: L356, F374, P375, L378, M379, I381 and A389 (yellow); | Hydrophobic residues: L356, F374, P375, L378, M379, I381 and A389 (yellow); | ||
Negatively charged residues: D338, D349, E353, E357, D383, and E393 (red); | Negatively charged residues: D338, D349, E353, E357, D383, and E393 (red); | ||
| Line 17: | Line 17: | ||
The residues of | The residues of <scene name='RA_Mediated_T-reg_Differentiaition/Rxr_dimer_interface/2'>RXR-</scene>α that are interacting in the heterodimer are as follows: | ||
Hydrophobic residues: Y402, P417, F420, A421, L424, L425, L427, P428, A429, and L435 (yellow); | Hydrophobic residues: Y402, P417, F420, A421, L424, L425, L427, P428, A429, and L435 (yellow); | ||
Negatively charged residues: E357, D384, E395, E399, E406, and E439 (red); | Negatively charged residues: E357, D384, E395, E399, E406, and E439 (red); | ||
Revision as of 09:56, 14 November 2012
IntroductionIntroduction
T-regulatory cells (T-regs) are a small subset of CD4+ T-cells that exhibit strong down regulation of immune system activity in their local environment. They are distinguished from other CD4+ T-cells by the expression of FOXP3, a gene regulator. [1] The exact mechanisms used by T-regs to down regulate the immune system has not yet been clearly elucidated. These cells have been shown to differentiate from CD4+ T-helper cells upon activation and exposure to the following cytokines: tumor growth factor β (TGF-β), Interleukin-2 (IL-2) and retinoic acid (RA). [2] Both TGF-β and IL-2 are used in other immune system differentiation, however, RA has been shown to bias T-cells to the T-reg phenotype. [3] When acting upon T-reg cells, RA acts as the ligand for the Retinoic Acid Receptor-α (RARα) / Retinoid X Receptor-α (RXRα) heterodimer. This heterodimer is of the nuclear receptor family, and each chain consists of the same three part structure: a Ligand binding domain (LBD), a DNA binding domain (DBD), and a hinge region connecting the two binding domains. [4]
| Ligand Binding DomainThe Ligand binding domain for each piece of the dimer has a nearly identical structure of an . These alpha helices form a total of 12 domains per protein (referred to as H1-12), with an additional 2 beta sheets as well. Both monomers contain two regions of activity, the and the ligand binding domain.[5]
RARα-RXRα HeterodimerWhen these two proteins form a heterodimer, the overall structure of the larger dimer is comparable to that of an RXRα homodimer, likely due to the many similarities these two molecules share. RARα and RXRα rely on residues from the H7, H8, H9, H10, L8-9, and L9-10 domains of both molecules to form the . The sequence identity between the two molecules on the dimer interface is 0.33, demonstrating that 33% of the interacting residues are homologous between the different proteins. [6]
The residues of α that are interacting in the heterodimer are as follows: Hydrophobic residues: L356, F374, P375, L378, M379, I381 and A389 (yellow); Negatively charged residues: D338, D349, E353, E357, D383, and E393 (red); Positively charged residues: K360, R364, H372, K376, K380, and R385 (blue); Hydrophilic residues: Q315, Q352, T382, and S386 (green).
The residues of α that are interacting in the heterodimer are as follows: Hydrophobic residues: Y402, P417, F420, A421, L424, L425, L427, P428, A429, and L435 (yellow); Negatively charged residues: E357, D384, E395, E399, E406, and E439 (red); Positively charged residues: R353, K361, R398, K410, K422, R426, R431, and K436 (blue); Hydrophilic residues: S432 (green).
RARα-RXRα Ligand Interactions | ||||||||||
DNA Binding DomainRXRα-DNA InteractionsDR-5 Specificity |
| ||||||||||
Biological SignificanceBiological Significance
ReferencesReferences
- ↑ Ochs HD, Oukka M, Torgerson TR. TH17 cells and regulatory T cells in primary immunodeficiency diseases. J Allergy Clin Immunol. 2009 May;123(5):977-83; quiz 984-5. PMID:19410687 doi:10.1016/j.jaci.2009.03.030
- ↑ Moore C, Fuentes C, Sauma D, Morales J, Bono MR, Rosemblatt M, Fierro JA. Retinoic acid generates regulatory T cells in experimental transplantation. Transplant Proc. 2011 Jul-Aug;43(6):2334-7. PMID:21839265 doi:10.1016/j.transproceed.2011.06.057
- ↑ Moore C, Fuentes C, Sauma D, Morales J, Bono MR, Rosemblatt M, Fierro JA. Retinoic acid generates regulatory T cells in experimental transplantation. Transplant Proc. 2011 Jul-Aug;43(6):2334-7. PMID:21839265 doi:10.1016/j.transproceed.2011.06.057
- ↑ Kumar R, Thompson EB. The structure of the nuclear hormone receptors. Steroids. 1999 May;64(5):310-9. PMID:10406480
- ↑ Bourguet W, Vivat V, Wurtz JM, Chambon P, Gronemeyer H, Moras D. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol Cell. 2000 Feb;5(2):289-98. PMID:10882070
- ↑ Bourguet W, Vivat V, Wurtz JM, Chambon P, Gronemeyer H, Moras D. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol Cell. 2000 Feb;5(2):289-98. PMID:10882070