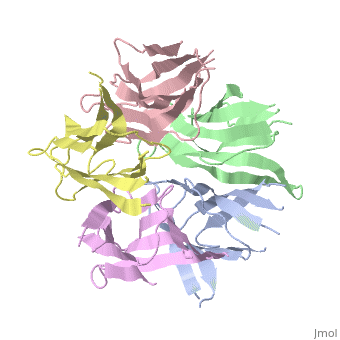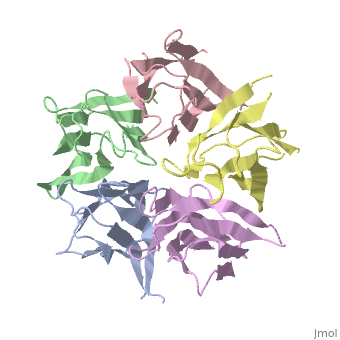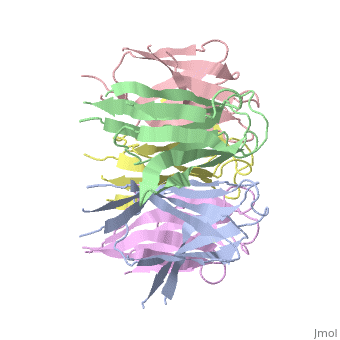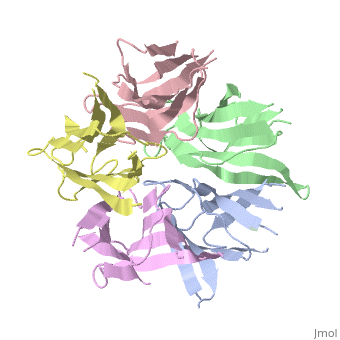1k5j: Difference between revisions
New page: left|200px<br /> <applet load="1k5j" size="450" color="white" frame="true" align="right" spinBox="true" caption="1k5j, resolution 2.30Å" /> '''The Crystal Structu... |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Image:1k5j.gif|left|200px]]<br /> | [[Image:1k5j.gif|left|200px]]<br /><applet load="1k5j" size="350" color="white" frame="true" align="right" spinBox="true" | ||
<applet load="1k5j" size=" | |||
caption="1k5j, resolution 2.30Å" /> | caption="1k5j, resolution 2.30Å" /> | ||
'''The Crystal Structure of Nucleoplasmin-Core'''<br /> | '''The Crystal Structure of Nucleoplasmin-Core'''<br /> | ||
==Overview== | ==Overview== | ||
The efficient assembly of histone complexes and nucleosomes requires the | The efficient assembly of histone complexes and nucleosomes requires the participation of molecular chaperones. Currently, there is a paucity of data on their mechanism of action. We now present the structure of an N-terminal domain of nucleoplasmin (Np-core) at 2.3 A resolution. The Np-core monomer is an eight-stranded beta barrel that fits snugly within a stable pentamer. In the crystal, two pentamers associate to form a decamer. We show that both Np and Np-core are competent to assemble large complexes that contain the four core histones. Further experiments and modeling suggest that these complexes each contain five histone octamers which dock to a central Np decamer. This work has important ramifications for models of histone storage, sperm chromatin decondensation, and nucleosome assembly. | ||
==About this Structure== | ==About this Structure== | ||
1K5J is a [http://en.wikipedia.org/wiki/Single_protein Single protein] structure of sequence from [http://en.wikipedia.org/wiki/Xenopus_laevis Xenopus laevis]. The following page contains interesting information on the relation of 1K5J with [[http://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/pdb85_1.html Importins]]. Full crystallographic information is available from [http:// | 1K5J is a [http://en.wikipedia.org/wiki/Single_protein Single protein] structure of sequence from [http://en.wikipedia.org/wiki/Xenopus_laevis Xenopus laevis]. The following page contains interesting information on the relation of 1K5J with [[http://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/pdb85_1.html Importins]]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1K5J OCA]. | ||
==Reference== | ==Reference== | ||
| Line 15: | Line 14: | ||
[[Category: Single protein]] | [[Category: Single protein]] | ||
[[Category: Xenopus laevis]] | [[Category: Xenopus laevis]] | ||
[[Category: Akey, C | [[Category: Akey, C W.]] | ||
[[Category: Akey, I | [[Category: Akey, I V.]] | ||
[[Category: Dingwall, C.]] | [[Category: Dingwall, C.]] | ||
[[Category: Dutta, S.]] | [[Category: Dutta, S.]] | ||
[[Category: Hartman, K | [[Category: Hartman, K L.]] | ||
[[Category: Head, J | [[Category: Head, J F.]] | ||
[[Category: Laue, T.]] | [[Category: Laue, T.]] | ||
[[Category: Nolte, R | [[Category: Nolte, R T.]] | ||
[[Category: beta-barrel]] | [[Category: beta-barrel]] | ||
[[Category: beta-bulge]] | [[Category: beta-bulge]] | ||
| Line 28: | Line 27: | ||
[[Category: pentamer]] | [[Category: pentamer]] | ||
''Page seeded by [http:// | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Feb 21 13:30:25 2008'' | ||
Revision as of 14:30, 21 February 2008
|
The Crystal Structure of Nucleoplasmin-Core
OverviewOverview
The efficient assembly of histone complexes and nucleosomes requires the participation of molecular chaperones. Currently, there is a paucity of data on their mechanism of action. We now present the structure of an N-terminal domain of nucleoplasmin (Np-core) at 2.3 A resolution. The Np-core monomer is an eight-stranded beta barrel that fits snugly within a stable pentamer. In the crystal, two pentamers associate to form a decamer. We show that both Np and Np-core are competent to assemble large complexes that contain the four core histones. Further experiments and modeling suggest that these complexes each contain five histone octamers which dock to a central Np decamer. This work has important ramifications for models of histone storage, sperm chromatin decondensation, and nucleosome assembly.
About this StructureAbout this Structure
1K5J is a Single protein structure of sequence from Xenopus laevis. The following page contains interesting information on the relation of 1K5J with [Importins]. Full crystallographic information is available from OCA.
ReferenceReference
The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly., Dutta S, Akey IV, Dingwall C, Hartman KL, Laue T, Nolte RT, Head JF, Akey CW, Mol Cell. 2001 Oct;8(4):841-53. PMID:11684019
Page seeded by OCA on Thu Feb 21 13:30:25 2008