1egp: Difference between revisions
New page: left|200px<br /><applet load="1egp" size="450" color="white" frame="true" align="right" spinBox="true" caption="1egp, resolution 2.0Å" /> '''PROTEINASE INHIBITOR ... |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Image:1egp.jpg|left|200px]]<br /><applet load="1egp" size=" | [[Image:1egp.jpg|left|200px]]<br /><applet load="1egp" size="350" color="white" frame="true" align="right" spinBox="true" | ||
caption="1egp, resolution 2.0Å" /> | caption="1egp, resolution 2.0Å" /> | ||
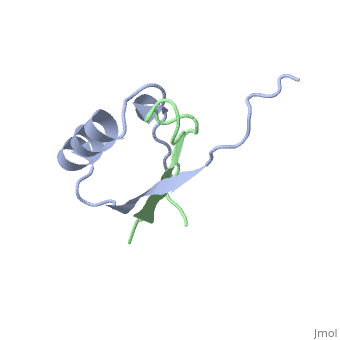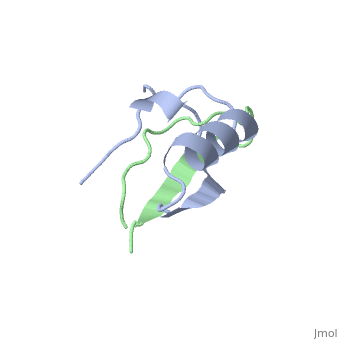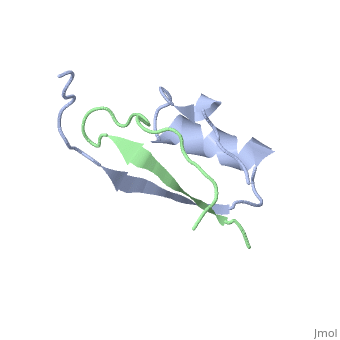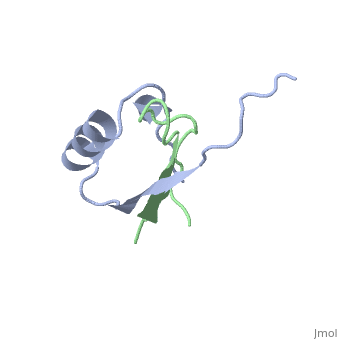
'''PROTEINASE INHIBITOR EGLIN C WITH HYDROLYSED REACTIVE CENTER'''<br /> | '''PROTEINASE INHIBITOR EGLIN C WITH HYDROLYSED REACTIVE CENTER'''<br /> | ||
==Overview== | ==Overview== | ||
The inhibition of serine proteinases by both synthetic and natural | The inhibition of serine proteinases by both synthetic and natural inhibitors has been widely studied. Eglin c is a small thermostable protein isolated from the leech, Hirudo medicinalis. Eglin c is a potent serine proteinase inhibitor. The three-dimensional structure of native eglin and of its complexes with a number of proteinases are known. We here describe the crystal structure of hydrolysed eglin not bound to a proteinase. The body of the eglin has a conformation remarkably similar to that in the known complexes with proteinases. However, the peptide chain has been cut at the 'scissile' bond between residues 45 and 46, presumed to result from the presence of subtilisin DY in the crystallisation sample. The residues usually making up the inhibiting loop of eglin take up a quite different conformation in the nicked inhibitor leading to stabilising contacts between neighbouring molecules in the crystal. The structure was solved by molecular replacement techniques and refined to a final R-factor of 14.5%. | ||
==About this Structure== | ==About this Structure== | ||
1EGP is a [http://en.wikipedia.org/wiki/Protein_complex Protein complex] structure of sequences from [http://en.wikipedia.org/wiki/Hirudo_medicinalis Hirudo medicinalis]. Full crystallographic information is available from [http:// | 1EGP is a [http://en.wikipedia.org/wiki/Protein_complex Protein complex] structure of sequences from [http://en.wikipedia.org/wiki/Hirudo_medicinalis Hirudo medicinalis]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1EGP OCA]. | ||
==Reference== | ==Reference== | ||
| Line 16: | Line 16: | ||
[[Category: Dauter, Z.]] | [[Category: Dauter, Z.]] | ||
[[Category: Lamzin, V.]] | [[Category: Lamzin, V.]] | ||
[[Category: Wilson, K | [[Category: Wilson, K S.]] | ||
[[Category: proteinase inhibitor]] | [[Category: proteinase inhibitor]] | ||
''Page seeded by [http:// | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Feb 21 12:27:34 2008'' | ||
Revision as of 13:27, 21 February 2008
|
PROTEINASE INHIBITOR EGLIN C WITH HYDROLYSED REACTIVE CENTER
OverviewOverview
The inhibition of serine proteinases by both synthetic and natural inhibitors has been widely studied. Eglin c is a small thermostable protein isolated from the leech, Hirudo medicinalis. Eglin c is a potent serine proteinase inhibitor. The three-dimensional structure of native eglin and of its complexes with a number of proteinases are known. We here describe the crystal structure of hydrolysed eglin not bound to a proteinase. The body of the eglin has a conformation remarkably similar to that in the known complexes with proteinases. However, the peptide chain has been cut at the 'scissile' bond between residues 45 and 46, presumed to result from the presence of subtilisin DY in the crystallisation sample. The residues usually making up the inhibiting loop of eglin take up a quite different conformation in the nicked inhibitor leading to stabilising contacts between neighbouring molecules in the crystal. The structure was solved by molecular replacement techniques and refined to a final R-factor of 14.5%.
About this StructureAbout this Structure
1EGP is a Protein complex structure of sequences from Hirudo medicinalis. Full crystallographic information is available from OCA.
ReferenceReference
Structure of the proteinase inhibitor eglin c with hydrolysed reactive centre at 2.0 A resolution., Betzel C, Dauter Z, Genov N, Lamzin V, Navaza J, Schnebli HP, Visanji M, Wilson KS, FEBS Lett. 1993 Feb 15;317(3):185-8. PMID:8425603
Page seeded by OCA on Thu Feb 21 12:27:34 2008