ToxT: Difference between revisions
No edit summary |
No edit summary |
||
| Line 7: | Line 7: | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
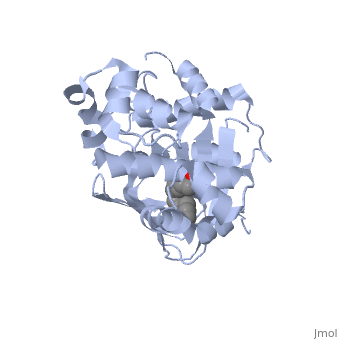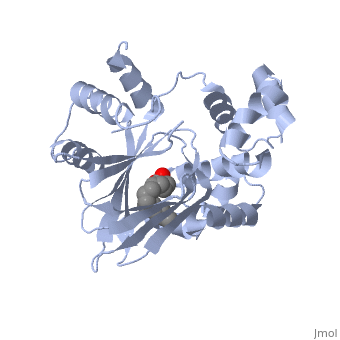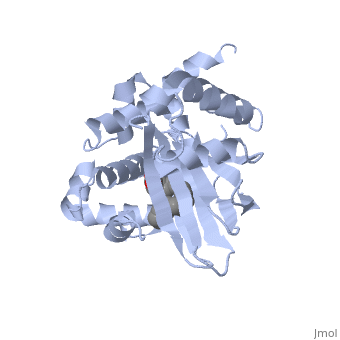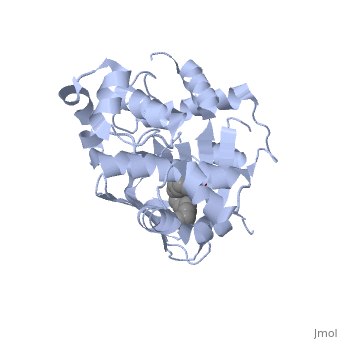
<u>Ligand-binding</u>. <scene name='ToxT_Transcriptional_Regulator_in_Vibrio_cholerae/Barrel/1'>A nine-stranded beta sheet sandwich</scene> or "jelly-roll" with three other alpha helices (overall making up the <scene name='ToxT_Transcriptional_Regulator_in_Vibrio_cholerae/N-terminus/1'>N-terminus</scene>) contain a <scene name='ToxT_Transcriptional_Regulator_in_Vibrio_cholerae/Binding_pocket/1'>binding pocket</scene>. This is made from several residues from the N-terminus (Y12, Y20, F22, L25, I27, K31, F33, L61, F69, L71, V81, and V83), and a few from the C-terminus (I226, K230, M259, V261, Y266, and M269). The pocket is highly hydrophobic, and has a known volume of 780.9 Angstroms.<ref name="structure">PMID: 20133655</ref> This pocket contains a ligand: <scene name='ToxT_Transcriptional_Regulator_in_Vibrio_cholerae/Binding_pocket/2'>cis-palmitoleate</scene> <ref name="structure">PMID: 20133655</ref> which appears to have a negative effect on virulence when present in vitro. The <i>cis</i>-palmitoleate forms <scene name='ToxT_Transcriptional_Regulator_in_Vibrio_cholerae/Salt_bridges_pam/1'>salt bridges</scene> with residues K31 and K230 (for detail, see Figure 1B of: [http://www.pnas.org/content/107/7/2860/F1.large.jpg]). This unsaturated fatty acid, like other UFAs,[http://en.wikipedia.org/wiki/Fatty_acid#Unsaturated_fatty_acids] tend to inhibit genes under the control of ToxT. | <u>Ligand-binding</u>. <scene name='ToxT_Transcriptional_Regulator_in_Vibrio_cholerae/Barrel/1'>A nine-stranded beta sheet sandwich</scene> or "jelly-roll" with three other alpha helices (overall making up the <scene name='ToxT_Transcriptional_Regulator_in_Vibrio_cholerae/N-terminus/1'>N-terminus</scene>) contain a <scene name='ToxT_Transcriptional_Regulator_in_Vibrio_cholerae/Binding_pocket/1'>binding pocket</scene>. This is made from several residues from the N-terminus (Y12, Y20, F22, L25, I27, K31, F33, L61, F69, L71, V81, and V83), and a few from the C-terminus (I226, K230, M259, V261, Y266, and M269). This pocket contains a sixteen-carbon fatty acid positioned in a conformation such that its negatively charged carboxylate group forms salt bridges between K31 of the N-terminal domain, and K230 from the C-terminal domain. The pocket is highly hydrophobic, and has a known volume of 780.9 Angstroms.<ref name="structure">PMID: 20133655</ref> This pocket contains a ligand: <scene name='ToxT_Transcriptional_Regulator_in_Vibrio_cholerae/Binding_pocket/2'>cis-palmitoleate</scene> <ref name="structure">PMID: 20133655</ref> which appears to have a negative effect on virulence when present in vitro. The <i>cis</i>-palmitoleate forms <scene name='ToxT_Transcriptional_Regulator_in_Vibrio_cholerae/Salt_bridges_pam/1'>salt bridges</scene> with residues K31 and K230 (for detail, see Figure 1B of: [http://www.pnas.org/content/107/7/2860/F1.large.jpg]). This unsaturated fatty acid, like other UFAs,[http://en.wikipedia.org/wiki/Fatty_acid#Unsaturated_fatty_acids] tend to inhibit genes under the control of ToxT. | ||
Specifically, the <i>cis</i>-palmitoleate (PAM) appears to change ToxT's conformation, and thus lower its ability to bind DNA and form dimers.<ref name="structure">PMID: 20133655</ref> The presence of UFAs is associated with being in the lumen of the intestine during the bacterial infection. PAM brings K31 and K230 together from either end of the protein, and essentially closes off ToxT. K230 is at the end of helix seven, and binding to K31 causes helix six to be pulled into an unfavorable conformation that deters DNA binding. In lower concentration of fatty acids, ie: after penetrating the intestine's mucus, PAM is in lower concentration and tends subsequently not to be bound, and the two lysine residues repel each other to give ToxT an open conformation. The freedom of helices six and seven to find a favorable configuration allows DNA binding to occur.<ref name="structure">PMID: 20133655</ref> | Specifically, the <i>cis</i>-palmitoleate (PAM) appears to change ToxT's conformation, and thus lower its ability to bind DNA and form dimers.<ref name="structure">PMID: 20133655</ref> The presence of UFAs is associated with being in the lumen of the intestine during the bacterial infection. PAM brings K31 and K230 together from either end of the protein, and essentially closes off ToxT. K230 is at the end of helix seven, and binding to K31 causes helix six to be pulled into an unfavorable conformation that deters DNA binding. In lower concentration of fatty acids, ie: after penetrating the intestine's mucus, PAM is in lower concentration and tends subsequently not to be bound, and the two lysine residues repel each other to give ToxT an open conformation. The freedom of helices six and seven to find a favorable configuration allows DNA binding to occur.<ref name="structure">PMID: 20133655</ref> | ||
<br/> | <br/> | ||
Revision as of 21:59, 4 May 2012
The crystal structure of ToxT is resolved in monomeric form, after isolation from Vibrio cholerae strain O395.[1]
IntroductionIntroduction
ToxT is a molecule at the end of a transcriptional cascade that autoregulates the transcription of the primary virulence factors of Vibrio cholerae[3] and itself. ToxT is a cytoplasmic protein that is activated in turn by ToxR, which is itself activated by ToxS in response to environmental stimuli.[2] These two factors, cholera toxin (CT)[4] and the toxin co-regulated pilus (TCP), are instrumental in causing the disease cholera[5]. This is an intestinal infection resulting in massive water loss in the affected individual, causing extreme dehydration.[6] Rehydration is sufficient as treatment.
Structural FeaturesDNA-binding. ToxT belongs to a family of transcriptional regulators headed by and known as AraC.[1] The AraC family is characterized by a 100 amino acid region of sequence similarity that forms a with two helix-turn-helix motifs (one on either side of the black linker). [3] This DNA binding domain is composed of seven alpha helices. HTH1 is composed of alpha helices five and six, while HTH2 is composed of alpha helices eight and nine. The two HTH regions are linked by the very polar alpha helix seven(shown in black). The overall domain is located at the C-terminus.[1] Assuming ToxT is similar in mechanism to other AraC proteins, helix six from HTH1 and helix nine from HTH2 become aligned with the help of helix seven. Helix seven is positioned to attach to the N terminal binding pocket(the polar linking region) to allow binding to major consecutive grooves of target DNA (specific promoters for virulence genes).[1][1]. The conformation of helix seven is dependent on the ligand bound.
LigandIn this resolved structure, [5] is shown, which can be bound in the beta sheet barrel (as discussed above). This unsaturated fatty acid reduces virulence expression in Vibrio cholerae. |
| ||||||||||
Further StudyFurther Study
Conclusive results about what activates ToxT itself has not yet been found. The varying activity of ToxT dependent on the presence of cis-palmitoleate or other unsaturated fatty acids represents a detailed method of effective pathogenicity in humans, but may not be a reasonable target for drug treatment. By restricting transcription (and thus translation and protein production) of virulence genes until the bacterium is determined to be in a favorable location for infection, Vibrio cholerae avoids wasting energy producing virulence factors that will just be cleared by the intestine. This is a specific mechanism to ensure that the bacterium also injects CT and TCP where they will do the most damage, perpetuating the infection. [6]
ReferencesReferences
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Lowden MJ, Skorupski K, Pellegrini M, Chiorazzo MG, Taylor RK, Kull FJ. Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc Natl Acad Sci U S A. 2010 Feb 16;107(7):2860-5. Epub 2010 Feb 1. PMID:20133655
- ↑ Kenneth Todar [1] Vibrio cholerae and Asiatic Cholera, Todar's Online Textbook of Bacteriology. Date of access: 2011-11-28.
- ↑ Martin RG, Rosner JL. The AraC transcriptional activators. Curr Opin Microbiol. 2001 Apr;4(2):132-7. PMID:11282467
- ↑ Weber GG, Klose KE. The complexity of ToxT-dependent transcription in Vibrio cholerae. Indian J Med Res. 2011 Feb;133(2):201-6. PMID:21415495
- ↑ Shakhnovich EA, Hung DT, Pierson E, Lee K, Mekalanos JJ. Virstatin inhibits dimerization of the transcriptional activator ToxT. Proc Natl Acad Sci U S A. 2007 Feb 13;104(7):2372-7. Epub 2007 Feb 5. PMID:17283330 doi:10.1073/pnas.0611643104
- ↑ Kenneth Todar [2] Vibrio cholerae and Asiatic Cholera, Todar's Online Textbook of Bacteriology. Date of access: 2011-11-28.