Amyloid beta: Difference between revisions
Laura Olney (talk | contribs) No edit summary |
Laura Olney (talk | contribs) No edit summary |
||
| Line 6: | Line 6: | ||
Amyoloid beta is actually the <scene name='Amyloid_beta/C-term/1'>C-terminal</scene> of the <scene name='Amyloid_beta/App/1'>Amyloid Precursor Protein</scene> which is a type I membrane-spanning glycoprotein encoded on chromosome 21 [[http://proteopedia.org/wiki/index.php/Amyloid_precursor_protein APP]].<ref name="alz" /> Amyloid beta results from an abnormal cleavage by [http://proteopedia.org/wiki/index.php/Beta_secretase beta-secretase] at the N-terminal and gamma-secretase at the C-terminal[[http://en.wikipedia.org/wiki/Beta_amyloid]]. The cleavage is nonspecific and results in peptides 39-43 amino acids in length, with 42 being the most common. Such cleavages occur most commonly in the plasma membrane though it can also occur in neuronal membranes.<ref name="alz" /> | Amyoloid beta is actually the <scene name='Amyloid_beta/C-term/1'>C-terminal</scene> of the <scene name='Amyloid_beta/App/1'>Amyloid Precursor Protein</scene> which is a type I membrane-spanning glycoprotein encoded on chromosome 21 [[http://proteopedia.org/wiki/index.php/Amyloid_precursor_protein APP]].<ref name="alz" /> Amyloid beta results from an abnormal cleavage by [http://proteopedia.org/wiki/index.php/Beta_secretase beta-secretase] at the N-terminal and gamma-secretase at the C-terminal[[http://en.wikipedia.org/wiki/Beta_amyloid]]. The cleavage is nonspecific and results in peptides 39-43 amino acids in length, with 42 being the most common. Such cleavages occur most commonly in the plasma membrane though it can also occur in neuronal membranes.<ref name="alz" /> | ||
The first 16 residues, Asp-Ala-Glu-Phe-Arg-His-Asp-Ser-Gly-Tyr-Glu--Val-His-His-Gln-Lys, are mostly hydrophobic with <scene name='Amyloid_beta/Cu/1'>His13 and His14</scene> | The first 16 residues, Asp-Ala-Glu-Phe-Arg-His-Asp-Ser-Gly-Tyr-Glu--Val-His-His-Gln-Lys, are mostly hydrophobic with <scene name='Amyloid_beta/Cu/1'>His13 and His14</scene> acting as a binding domain for Cu(II). Residues <scene name='Amyloid_beta/Self/2'>12-23</scene> function as the self recognition region allowing for the formation of dimers and/or oligomers. This region also serves as the binding site for cholesterol, apolipoproteinE, alpha7nAChr, and amyloid beta-peptide binding alcohol dehydrogenase.<ref name="alz" /> | ||
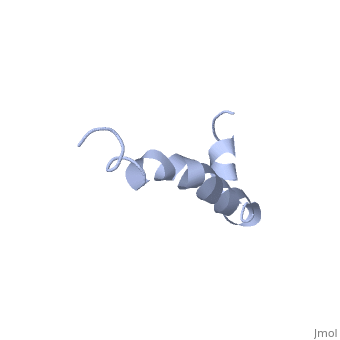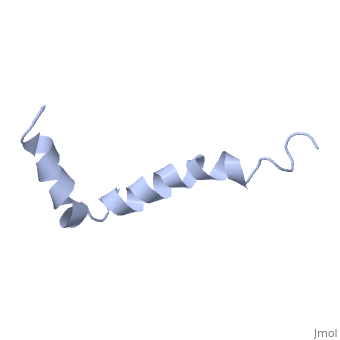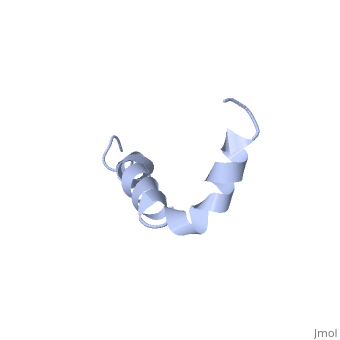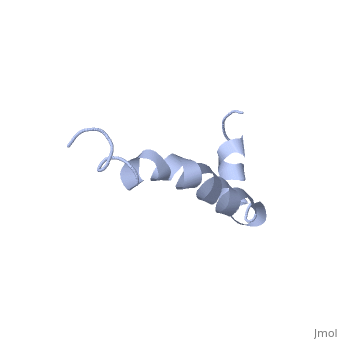
The most reasonable structure determined structure consists of <scene name='Amyloid_beta/Structure/1'>two helices</scene>; the first helix (residues 8-25) is well defined and has an RMSD of 0.38 angstroms and the second (residues 28-38) is interrupted at the Ile32-Gly33 connection. The second helix corresponds to the transmembrane region of APP and thus contains multiple small and hydrophobic amino acids. The two helices are connected by a <scene name='Amyloid_beta/Kink/1'>kink</scene> (residues 26 and 27).<ref name="structure" /> | The most reasonable structure determined structure consists of <scene name='Amyloid_beta/Structure/1'>two helices</scene>; the first helix (residues 8-25) is well defined and has an RMSD of 0.38 angstroms and the second (residues 28-38) is interrupted at the Ile32-Gly33 connection. The second helix corresponds to the transmembrane region of APP and thus contains multiple small and hydrophobic amino acids. The two helices are connected by a <scene name='Amyloid_beta/Kink/1'>kink</scene> (residues 26 and 27).<ref name="structure" /> | ||
Revision as of 22:05, 29 November 2011
IntroductionAlzheimer's disease is characterized by extracellular proteic plaques and intracellular neurofibril tangles.[1] These plaques are collections of beta-amyloid of beta-sheets. The fibrils form when normally soluble amyloid beta proteins reach a critical concentration and become insoluble, misfold, and aggregate.[2] When amyloid beta comes into contact with metal ions and oxygen the result is the production of reactive oxygen species (especially hydrogen peroxide) without oxygen and metal ions amyloid beta ca induce pore formation in neuronal and endothelial cells, triggering cell death.[1][3] Yet another source of amyloid beta toxicity stems from its ability to induce endothelial cell damage through the production of superoxide, though the mechanism of such induction is unclear.[3] While the presence of the fibril plaques remains a marker for Alzheimer's disease recent studies have suggested that alymoild beta oligomers most devestating effect is the impairment of long-term potentiation which decreases dendritic spine density in the hippocampal brain and impairs memory.[4] StructureAmyoloid beta is actually the of the which is a type I membrane-spanning glycoprotein encoded on chromosome 21 [APP].[3] Amyloid beta results from an abnormal cleavage by beta-secretase at the N-terminal and gamma-secretase at the C-terminal[[1]]. The cleavage is nonspecific and results in peptides 39-43 amino acids in length, with 42 being the most common. Such cleavages occur most commonly in the plasma membrane though it can also occur in neuronal membranes.[3] The first 16 residues, Asp-Ala-Glu-Phe-Arg-His-Asp-Ser-Gly-Tyr-Glu--Val-His-His-Gln-Lys, are mostly hydrophobic with acting as a binding domain for Cu(II). Residues function as the self recognition region allowing for the formation of dimers and/or oligomers. This region also serves as the binding site for cholesterol, apolipoproteinE, alpha7nAChr, and amyloid beta-peptide binding alcohol dehydrogenase.[3] The most reasonable structure determined structure consists of ; the first helix (residues 8-25) is well defined and has an RMSD of 0.38 angstroms and the second (residues 28-38) is interrupted at the Ile32-Gly33 connection. The second helix corresponds to the transmembrane region of APP and thus contains multiple small and hydrophobic amino acids. The two helices are connected by a (residues 26 and 27).[1] The toxicity of amyloid beta is due to the formation of aggregates and oligomers. The actual structures of these oligomers is unknown but is likely similar to that of amyloid beta in mature plaques.[1]
|
| ||||||||||
NeurotoxicityNeurotoxicity
Generation of Radicals Amyloid beta produces reactive oxygen species, which induces oxidative stress and inflammation.[3]
Pore Formation Amyloid beta can adhere to endothelial cell walls and create lesions, eventually a large deposite will cause cerrbral hemorrhage. The pores cause loss of calcium homeostasis and an influx of Ca2+ into neurons.[3]
Interaction with tau Tau proteins function to stabilize microtubules [[1]]. Association between Tau proteins and amyloid beta results in dissociation of tau from microtubules which collapses the axonal structure leading to the death of neurons.[3]
Binding ApoE, ABAD, and catalase By binding these proteins amyloid beta alters the normal activities and leads to over production of amyloid beta and oxidative stress.
PreventionPrevention
The most promising prevention of amyloid beta has been to prevent the enzymes responsible for its creation. AF267B which is a muscarine receptor that activates aplha-secretase and reduces tau pathology.[3] Thus far no true treatment or prevention has been determined for Alzheimer's.
ReferencesReferences
- ↑ 1.0 1.1 1.2 1.3 Crescenzi O, Tomaselli S, Guerrini R, Salvadori S, D'Ursi AM, Temussi PA, Picone D. Solution structure of the Alzheimer amyloid beta-peptide (1-42) in an apolar microenvironment. Similarity with a virus fusion domain. Eur J Biochem. 2002 Nov;269(22):5642-8. PMID:12423364
- ↑ Vlassenko AG, Benzinger TL, Morris JC. PET amyloid-beta imaging in preclinical Alzheimer's disease. Biochim Biophys Acta. 2011 Nov 12. PMID:22108203 doi:10.1016/j.bbadis.2011.11.005
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Cite error: Invalid
<ref>tag; no text was provided for refs namedalz - ↑ Funke SA. Detection of Soluble Amyloid-beta Oligomers and Insoluble High-Molecular-Weight Particles in CSF: Development of Methods with Potential for Diagnosis and Therapy Monitoring of Alzheimer's Disease. Int J Alzheimers Dis. 2011;2011:151645. Epub 2011 Nov 2. PMID:22114742 doi:10.4061/2011/151645