The Bacterial Flagellar Hook: Difference between revisions
Eric Martz (talk | contribs) |
Eric Martz (talk | contribs) |
||
| Line 20: | Line 20: | ||
== Hook Filament Structure == | == Hook Filament Structure == | ||
<table align="right" width="200" ><tr><td rowspan="2"> </td><td>[[Image:Samatey hook copyright nature 2004.gif]]</td></tr><tr><td>Bacterial flagellar hook, rotating simulation, used with permission of [[Fadel A. Samatey Group|Fadel Samatey]].</td></tr></table> | |||
In the original monomer structure paper<ref name="hook1" />, the monomer was fitted into an electron density map of about 10 Å resolution, generated by electron cryomicroscopy from hook filaments of ''Salmonella typhimurium''. Hook filaments are normally curved (at room temperature), but these hook filaments were rendered straight by cold storage, which facilitated image averaging. The D1 and D2 domains of the monomer were docked into the map and ligated. A model of the curved hook filament was generated using symmetry operations that produced the known radius of curvature. This model was then rotated, producing the simulation shown at right. | In the original monomer structure paper<ref name="hook1" />, the monomer was fitted into an electron density map of about 10 Å resolution, generated by electron cryomicroscopy from hook filaments of ''Salmonella typhimurium''. Hook filaments are normally curved (at room temperature), but these hook filaments were rendered straight by cold storage, which facilitated image averaging. The D1 and D2 domains of the monomer were docked into the map and ligated. A model of the curved hook filament was generated using symmetry operations that produced the known radius of curvature. This model was then rotated, producing the simulation shown at right. | ||
Revision as of 10:22, 25 May 2011
The bacterial flagellar hook described in this article is one part of the bacterial flagellum. Please see Flagella, bacterial (under development at Sandbox4 Eric Martz) for an overview of where the hook fits in the flagellum.
The flagellar hook is a molecular universal joint that transmits torque from the motor, anchored in the bacterial cell wall, to the flagellar filament, the relatively rigid helical rod that propels the bacterial cell when rotated. The hook is flexible: it enables the filament to adopt a wide range of angles relative to the motor axis and cell wall, yet continue to be rotated by the motor at all these angles.
Structure of the Hook Monomer, FlgEStructure of the Hook Monomer, FlgE
The hook filament of Salmonella typhimurium is composed of about 120 copies of the protein chain FlgE (the product of the gene flgE), a protein of length 402 amino acids.
FlgE31 2004FlgE31 2004
|
In 2004, Samatey et al. solved the structure of the mid-portion of the wild-type FlgE of Salmonella typhimurium by X-ray crystallography at a resolution of 1.8 Å (1wlg)[1]. The fragment successfully crystallized, designated FlgE31, consisted of amino acids 71-369, of which 71-363 were resolved (72% of the full-length 402-residue protein). Removal of the ends of the full-length chain (domain D0, see below) was required in order to coax the protein to crystallize, instead of forming filaments.
FlgE consists of three domains, D0, D1, and D2. The () at right shows D1 and D2 (D0 having been deleted in order to achieve crystallization, as mentioned above).
| Amino Terminus | Carboxy Terminus |
Note that D2 consists of a continuous segment of the protein chain, while D1 is made up of discontinuous segments. D0 (not shown, see above, also discontinuous) is attached to D1, and D0 forms the inner channel of the hook. D2 forms the outer surface of the hook.
[Show evolutionary conservation here after ConSurf server is again available.]
Hook Filament StructureHook Filament Structure
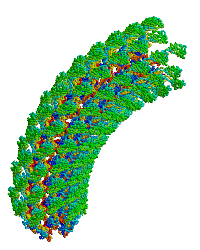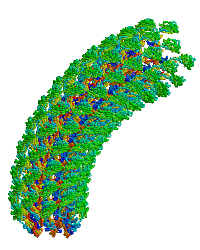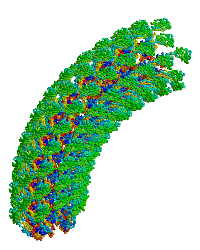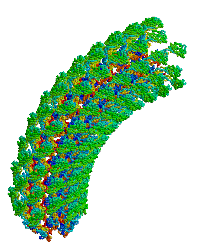 | |
| Bacterial flagellar hook, rotating simulation, used with permission of Fadel Samatey. |
In the original monomer structure paper[1], the monomer was fitted into an electron density map of about 10 Å resolution, generated by electron cryomicroscopy from hook filaments of Salmonella typhimurium. Hook filaments are normally curved (at room temperature), but these hook filaments were rendered straight by cold storage, which facilitated image averaging. The D1 and D2 domains of the monomer were docked into the map and ligated. A model of the curved hook filament was generated using symmetry operations that produced the known radius of curvature. This model was then rotated, producing the simulation shown at right.
Monomer Within the HookMonomer Within the Hook
From the curved model of the hook (see below), here is a short segment just . Each color represents one of the 11 protofilaments. Since each protofilament is 3 monomers in length, there are 33 monomers in this model. You can see how the Flg31 monomers fit into the hook when .
The monomers are slightly larger and more tightly packed than indicated in this model, because this model contains only alpha-carbon atoms.
Content AttributionContent Attribution
The initial content for this article was adapted, with permission, from The Bacterial Flagellar Hook: A Molecular Universal Joint, authored by User:Eric Martz in 2004-2006 for Protein Explorer.
References and NotesReferences and Notes
- ↑ 1.0 1.1 Samatey FA, Matsunami H, Imada K, Nagashima S, Shaikh TR, Thomas DR, Chen JZ, Derosier DJ, Kitao A, Namba K. Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism. Nature. 2004 Oct 28;431(7012):1062-8. PMID:15510139 doi:http://dx.doi.org/10.1038/nature02997