Bawel sandbox1: Difference between revisions
Seth Bawel (talk | contribs) No edit summary |
Seth Bawel (talk | contribs) No edit summary |
||
| Line 33: | Line 33: | ||
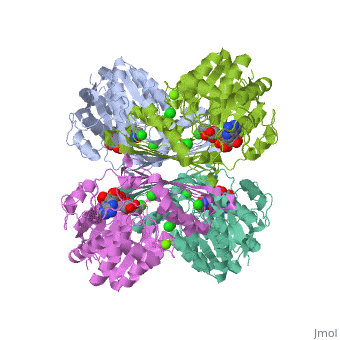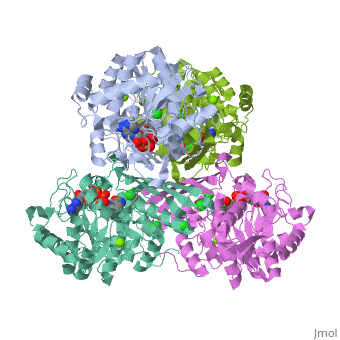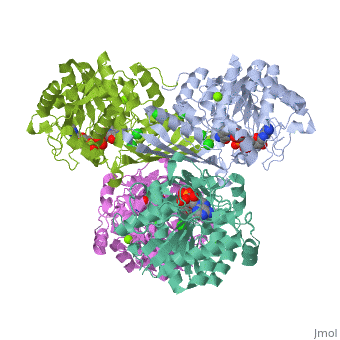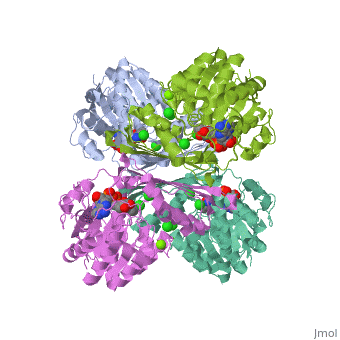
Glucose forms hydrogen bonds at the bottom of the deep crevice between the large domain and the small domain at the glucose binding site (active form). E256, E290 (shown in green) of the large domain, T168, K169 (shown in red) of the small domain, and N204, D205 (shown in yellow) of a connecting region form hydrogen bonds with glucose. The glucose binding site (inactive form) has a different conformation. At the allosteric site (active form), ATP forms hydrogen bonds with R63 and Y215 (shown in orange) and hydrophobically interacts with M210, Y214 (shown in blue) of the α5 helix and V452, V455 (shown in green) of the α13 helix. The allosteric site (inactive form) again shows structural differences. The differences in these two conformations allows glucokinase to function properly in different levels of glucose concentration. | |||
{{STRUCTURE_3cin | PDB=3cin | SCENE= }} | {{STRUCTURE_3cin | PDB=3cin | SCENE= }} | ||
Revision as of 07:28, 18 April 2011
hexokinase is an enzyme that phosphorylates a six-carbon sugar, a hexose, to a hexose phosphate. In most tissues and organisms, glucose is the most important substrate of hexokinases, and glucose 6-phosphate the most important product. Hexokinases have been found in every organism checked, ranging from bacteria, yeast, and plants, to humans and other vertebrates. They are categorized as actin fold proteins, sharing a common ATP binding site core surrounded by more variable sequences that determine substrate affinities and other properties. Several hexokinase isoforms or isozymes providing different functions can occur in a single species.
-Hexokinase I/A is found in all mammalian tissues, and is considered a "housekeeping enzyme," unaffected by most physiological, hormonal, and metabolic changes.
-Hexokinase II/B constitutes the principal regulated isoform in many cell types and is increased in many cancers.
-Hexokinase III/C is substrate-inhibited by glucose at physiologic concentrations. Little is known about the regulatory characteristics of this isoform.
-Hexokinase IV/D is also known as glucokinase and is described below.
Hexokinase Structure: The tertiary structure of hexokinase includes an open alpha/beta sheet. There is a large amount of variation associated with this structure. The ATP-binding domain is composed of five beta sheets and three alpha helices. In this open alph/beta sheet four of the beta sheets are parallel and one is in the anitparallel directions. The alpha helices and beta loops connect the beta sheets to produce this open alpha/beta sheet. The crevice indicates the ATP-binding domain of this glycolytic enzyme. The molecular weights of hexokinases are around 100 kD. Each consists of two similar 50kD halves, but only in hexokinase II do both halves have functional active sites.
Mechanism of Hexokinase: In the first reaction of glycolysis, the gamma-phosphoryl group of an ATP molecule is transferred to the oxygen at the C-6 of glucose. Hexokinase catalyzes this phosphoryl group transfer. To start this reaction, ATP forms a complex with magnesium (II) ion and glucose binds to kexokinase. The magnesium-ATP complex then binds with the hexokinase-glucose complex and forms an intermediate (Zeng, et al. present a picture showing the interctions of brain hexokinase with ATP). The hydroxyl group on the terminal phosphoryl group of the ATP molecule nucleophilically attacks the carbon 6 on glucose. This produces glucose-6-phosphate still bound to hexokinase and ADP still in complex with magnesium ion [5]. Glucose-6-phosphate and the magnesium-ADP complex leave hexokinase. Glucose-6-phosphate and ADP are the products of this reaction. Hexokinase undergoes an induced-fit conformational change when it binds to glucose, which ultimately prevents the hydrolysis of ATP. It also experiences potent allosteric inhibition under physiological concentrations by its immediate products, glucose-6-phosphate [4]. This is a mechanism by which the influx of substrate into the glycolytic pathway is controlled.
[edit] Glucokinase, an Isoenzyme of Hexokinase: Glucokinase (hexokinase D) is a monomeric cytoplasmic enzyme. As an isoenzyme of hexokinase, glucokinase is capable of prompting the first step of glycogen synthesis and glycolysis: the phosphorylation of glucose to glucose-6-phosphate (G6P). Since the phosphorylation of glucose to glucose-6-phosphate is the rate limiting step of glucose metabolism, glucokinase has a very important job in regulating healthy glucose levels in the human body [7].Mainly found in the liver and pancreas, glucokinase can also be found in the gut and brain.
Glucokinase vs. Other Hexokinases: Glucokinase is unique from other hexokinase in its kinetic properties and it is coded by a different gene. Compared to other hexokinases, glucokinase has a lower affinity, thus a higher Km, for glucose. Glucokinase actively stores glucose when serum glucose levels are high. Tissues lacking glucokinase use glucose at lower serum levels, so they use hexokinase with higher affinity for glucose. Also, G6P inhibits hexokinase (this is simple feedback inhibition). If the cell is not using up the G6P that it is making, then it should stop making it. However, G6P does not inhibit glucokinase. In fact, the steady state velocity of glucose phosphorylation displays a sigmoidal response curve to increasing glucose concentrations [7]. Glucokinase responds quickly to changes in glucose concentration becuase of these unique kinetic proeprties.
Structure of Glucokinase
Glucokinase Structure: Glucokinase also contains inactive and active conformations. Glucokinase consists of one chain or subunit of 448 amino acids forming a monomeric molecule consisting of 13 alpha helices and 5 beta sheets that can phosporylate glucose and other hexoses. The chain is folded into two distinct regions, a small and large domain. Glucokinase has one active binding site for glucose and one for ATP, which is the energy source for phosphorylation. This active binding site is located between the small and large domains. The carboxyl terminus is part of the alpha 13 helix, which codes for the region that forms half of the binding site for glucose. Glucokinase can be modulated to form an inactive and active complex. The inactive conformation forms when the alpha 13 helix has been modulated away from the rest of the molecule forming a large space. This space is too large to bind glucose so it is said to be in the inactive form. The alternative is when the alpha 13 helix is modulated to form a smaller space thus activating the protein[2].
Glucose forms hydrogen bonds at the bottom of the deep crevice between the large domain and the small domain at the glucose binding site (active form). E256, E290 (shown in green) of the large domain, T168, K169 (shown in red) of the small domain, and N204, D205 (shown in yellow) of a connecting region form hydrogen bonds with glucose. The glucose binding site (inactive form) has a different conformation. At the allosteric site (active form), ATP forms hydrogen bonds with R63 and Y215 (shown in orange) and hydrophobically interacts with M210, Y214 (shown in blue) of the α5 helix and V452, V455 (shown in green) of the α13 helix. The allosteric site (inactive form) again shows structural differences. The differences in these two conformations allows glucokinase to function properly in different levels of glucose concentration.
| |||||||||
| 3cin, resolution 1.70Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Gene: | TM1419, TM_1419 (Thermotoga maritima MSB8) | ||||||||
| Activity: | Inositol-3-phosphate synthase, with EC number 5.5.1.4 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum, TOPSAN | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Role in Organ Systems: In the liver glucokinase increases the synthesis of glycogen and is the first step in glycolysis, the main producer of ATP in the body. Glucokinase is responsible for phospohorylating the majority of glucose in the liver and pancreas. Glucokinase only binds to and phosphorylates glucose when levels are higher than normal blood glucose level, allowing it to maintain constant glucose levels[2]. By phosphorylating glucose, glucokinase creates glucose 6-phosphate. Glucose 6-phosphate can then be used by the liver through the glycolytic pathway. Along with this process in the liver, glucokinase also facilitates glycogen synthesis. Through this the majority of the body's glucose is stored. Glucose 6-phosphate is also one of the starting materials of the TCA cycle which is responsible for the majority of ATP production in the body.
In the pancreas, a rise in glucose levels increases the activity of glucokinase causing an increase in glucose 6-phosphate. This causes the triggering of the beta cells to secret insulin[3]. Glucokinase is the first step in this reaction. Insulin then allows other cells in the body to take up glucose, actively lowering the glucose level. Around 200 mutations inactivating glucokinase have been identified in triggering diabetes mellitus and 5 mutations activating glucokinase have been identified in triggering hypoglycemia [6]. Clearly, glucose plays a crucial role in maintaining healthy sugar levels in humans. Studies looking at the structure of gluockinase reveal the presence of allosteric site. This site is only present after glucose has attached to glucokinase [6]. Certain small molecules bind to this allosteric site and increase the activity of glucokinase [6]. These small molecules can potentially be used as a treatment for diabetes.
Hexokinase I is thought to be the "pacemaker of glycolysis in brain tissue and red blood cells [4].
[edit] Additional Resources For additional information, see: Carbohydrate Metabolism
[edit] References
1.↑ Pollard-Knight D, Cornish-Bowden A. Mechanism of liver glucokinase. Mol Cell Biochem. 1982 Apr 30;44(2):71-80. PMID:7048063
2.↑ 2.0 2.1 Kamata K, Mitsuya M, Nishimura T, Eiki J, Nagata Y. Structural basis for allosteric regulation of the monomeric allosteric enzyme human glucokinase. Structure. 2004 Mar;12(3):429-38. PMID:15016359 doi:10.1016/j.str.2004.02.005
3.↑ Postic C, Shiota M, Magnuson MA. Cell-specific roles of glucokinase in glucose homeostasis. Recent Prog Horm Res. 2001;56:195-217. PMID:11237213
4.↑ Zeng C, Aleshin A, Hardie J, Harrison R, Fromm H. ATP-Binding site of Human Brain Hexokinase as Studied by Molecular Modeling and Site-Directed Mutagenesis. Biochem. 1996 Aug 6;35:13157-13164.
5.↑ hammes G, and Kochavi D. Studies of the Enzyme Hexokinase: Kinetic Inhibition by Products. Massachusetts Institute of Technology. 1961 Oct 5.
6.↑ Ralph E, Thomson J, Almaden J, Sun S. Glucose Modulation fo Glucokinase Activation by Small Molecules. Biochem. 2008 Feb 15;47:5028-5036.
7.↑ Pal P, and Miller B. Activating Mutations in the Human Glucokinase Gene Revealed by Genetic Selection. Biochem. 2008 Dec 3;48:814-816.
s is a placeholder==
This is a placeholder text to help you get started in
placing a Jmol applet on your page. At any time, click
"Show Preview" at the bottom of this page to see how it goes.
Replace the PDB id (use lowercase!) after the STRUCTURE_ and after PDB= to load and display another structure.