Sandbox Reserved 321: Difference between revisions
No edit summary |
No edit summary |
||
| Line 10: | Line 10: | ||
[[Image:Secondary Structure of inhA.png|thumb|left|Width200Height300|alt=Secondary Structure Succession of inhA. Secondary structure residues are ordered from blue to red.|Secondary structure succession inhA.]] | [[Image:Secondary Structure of inhA.png|thumb|left|Width200Height300|alt=Secondary Structure Succession of inhA. Secondary structure residues are ordered from blue to red.|Secondary structure succession inhA.]] | ||
__TOC__ | __TOC__ | ||
| Line 20: | Line 18: | ||
=Structure of InhA= | =Structure of InhA= | ||
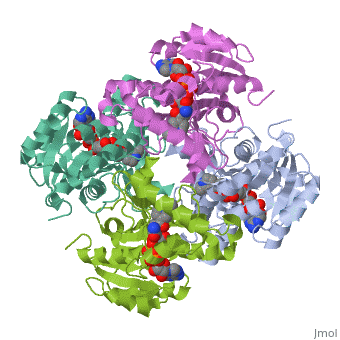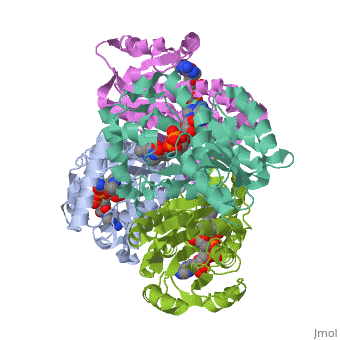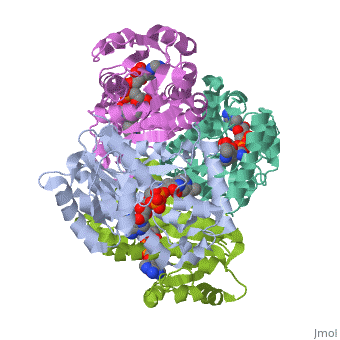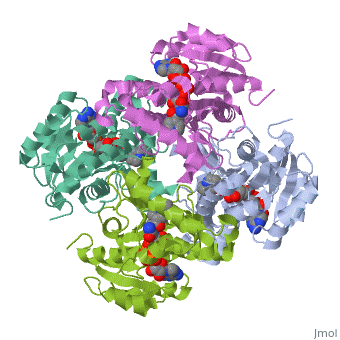
<Structure load='2h9i' size='275' frame='true' align='left' caption='Momomeric subunit of InhA with bound EAD' scene='Sandbox_Reserved_321/Structural_progresion/1' /> | |||
The inhA enzyme <scene name='Sandbox_Reserved_321/ | The inhA enzyme <scene name='Sandbox_Reserved_321/Structural_progresion/1'>(go to original scene)</scene> of ''M. tuberculosis'' is a homotetramer composed of a repeating subunit comprised of a single domain with a [http://en.wikipedia.org/wiki/Rossmann_fold Rossmann Fold] in the core that provides a NADH binding site<ref name ="crystallographic studies"/>. The single domain can be broken down into two substructures that are connected by short peptide loop<ref name ="making drugs for inhA"/><ref name ="crystallographic studies">PMID:17588773</ref>. The overall structure exhibits α/β folding of a series of α strands flanking a central β sheet of multiple parallel β strands<ref name ="crystallographic studies"/>. | ||
| Line 36: | Line 33: | ||
<scene name='Sandbox_Reserved_321/Substructure_2/1'>Substructure 2</scene> contains the c-terminal region of the molecule and consists of a small β strand <scene name='Sandbox_Reserved_321/B-7/1'>(B-7)</scene>, and two α helicies <scene name='Sandbox_Reserved_321/A-6_and_a-7/1'>(A-6 and A-7)</scene> which are conected by a short five residue loop<ref name ="making drugs for inhA"/>. The C-terminal domain consits of two other α helicies <scene name='Sandbox_Reserved_321/A-8_and_a-9/1'>(A-8 and A-9)</scene><ref name ="making drugs for inhA"/>. | <scene name='Sandbox_Reserved_321/Substructure_2/1'>Substructure 2</scene> contains the c-terminal region of the molecule and consists of a small β strand <scene name='Sandbox_Reserved_321/B-7/1'>(B-7)</scene>, and two α helicies <scene name='Sandbox_Reserved_321/A-6_and_a-7/1'>(A-6 and A-7)</scene> which are conected by a short five residue loop<ref name ="making drugs for inhA"/>. The C-terminal domain consits of two other α helicies <scene name='Sandbox_Reserved_321/A-8_and_a-9/1'>(A-8 and A-9)</scene><ref name ="making drugs for inhA"/>. | ||
| Line 51: | Line 42: | ||
The reaction takes place as follows. Intially NADH binds to the active site mediated by vander wall interactions with phenylalanine 41 (F41) and interations with lysine 165 <scene name='Sandbox_Reserved_321/Lys165/1'>(K165)</scene><ref name ="Roles of T158"/><ref name ="crystallographic studies"/>. Binding of NADH causes a conformational change in the Aspartate 42 and Arginine 43 <scene name='Sandbox_Reserved_321/Asp_42_and_arg_43/1'>(E42 and R43)</scene> side chains and an over all conformational change in InhA<ref name ="crystallographic studies"/>. In addition tyrosine 158 (Y158) playes an important role in alinging the carbonyl substrate, in fact; rotaion about its Cα-Cβ bond by 60° brings it into a position where it can hydrogen bond to the carbonyl of the 2-trans enoyl-ACP and provide it with electrophilic stabalization<ref name ="Roles of T158"/>. Inha then reduces the 2-trans double bond of the substrate by forming a enoyl intermediate through the transfer of a hydride ion from NADH to the thrid carbon of the substrate, followed by protonation of the second carbon<ref name ="crystallographic studies"/>. The biniding of both the substrate and the cofactor induces another conformational change in InhA that allows for the release of the meromycolic acid product<ref name ="crystallographic studies"/>. The meromycolic acids undergo | The reaction takes place as follows. Intially NADH binds to the active site mediated by vander wall interactions with phenylalanine 41 (F41) and interations with lysine 165 <scene name='Sandbox_Reserved_321/Lys165/1'>(K165)</scene><ref name ="Roles of T158"/><ref name ="crystallographic studies"/>. Binding of NADH causes a conformational change in the Aspartate 42 and Arginine 43 <scene name='Sandbox_Reserved_321/Asp_42_and_arg_43/1'>(E42 and R43)</scene> side chains and an over all conformational change in InhA<ref name ="crystallographic studies"/>. In addition tyrosine 158 (Y158) playes an important role in alinging the carbonyl substrate, in fact; rotaion about its Cα-Cβ bond by 60° brings it into a position where it can hydrogen bond to the carbonyl of the 2-trans enoyl-ACP and provide it with electrophilic stabalization<ref name ="Roles of T158"/>. Inha then reduces the 2-trans double bond of the substrate by forming a enoyl intermediate through the transfer of a hydride ion from NADH to the thrid carbon of the substrate, followed by protonation of the second carbon<ref name ="crystallographic studies"/>. The biniding of both the substrate and the cofactor induces another conformational change in InhA that allows for the release of the meromycolic acid product<ref name ="crystallographic studies"/>. The meromycolic acids undergo [http://en.wikipedia.org/wiki/Claisen_condensation claisen condensation] with a C26 fatty acid followed by reduction to a mature mycolic acid<ref name ="Fatty Acid Synthesis"/><ref name ="crystallographic studies"/>. | ||
=InhA and Thioamide Drugs= | =InhA and Thioamide Drugs= | ||
| Line 59: | Line 49: | ||
=Protein Superfamilly= | =Protein Superfamilly= | ||
InhA can also be classified into a family of short chain dehydrogenase/reductases (SDR). This family consists of proteins exhibiting a central core with a Rossmann fold that contains a NADH binding site. Examples of proteins in this family are listed below. | InhA can also be classified into a family of short chain dehydrogenase/reductases (SDR). This family consists of proteins exhibiting a central core with a Rossmann fold that contains a NADH binding site. There are appoximatley 3000 primary strucures outlines in various sequence databases<ref name ="SDR">PMID:12604210</ref>. Examples of proteins in this family are listed below. | ||
*[[1bxk]] - DTDP-glucose 4,6-dehydratase -''E. coli'' | |||
*[[1bsv]] - GDP-fructose synthetase in complex with NADPH - ''E. coli'' | |||
*[[1qrr]] - SQD1 | |||
*[[1ae1]] - Tyopinone reductase-I | |||
*[[ | |||
*[[ | |||
*[[ | |||
*[[ | |||
*[[ | |||
*[[ | |||
InhA can be further claasified into the acyl carrier protein family. These proteins generally all function in the transport of substrates in a myrid of pathways, such as: the sysnthesis of polyketides and fatty acids<ref name ="Acyl Carrier Proteins">PMID:17012233</ref>. Some examples of such proteins are listed below with links to their cooresponding proteopedia page. | InhA can be further claasified into the acyl carrier protein family. These proteins generally all function in the transport of substrates in a myrid of pathways, such as: the sysnthesis of polyketides and fatty acids<ref name ="Acyl Carrier Proteins">PMID:17012233</ref>. Some examples of such proteins are listed below with links to their cooresponding proteopedia page. | ||
Revision as of 09:42, 3 April 2011
| This Sandbox is Reserved from January 10, 2010, through April 10, 2011 for use in BCMB 307-Proteins course taught by Andrea Gorrell at the University of Northern British Columbia, Prince George, BC, Canada. |
To get started:
More help: Help:Editing |
InhAInhA
by Kelly Hrywkiw
| |||||||||
| 2h9i, resolution 2.20Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Gene: | inhA (Mycobacterium tuberculosis) | ||||||||
| Activity: | [acyl-carrier-protein_reductase_(NADH) Enoyl-[acyl-carrier-protein] reductase (NADH)], with EC number 1.3.1.9 | ||||||||
| Related: | 1zid | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
IntroductionIntroduction
The enzyme InhA is coded from the INHA gene that is simillar in sequence to the Salmonella typhimuriumgene which plays a role in fatty acid synthesis, and is part of a short chain dehydrogenase/reductase family[1][2]. Inha is an NADH dependent trans enoyl-acyl ACP carrier protein that is part of the fatty acid biosyntesis system: fatty acid synthase two (FASII), and plays a role in the sysnthesis of Mycolic Acid[3][2]. Mycolic acids are long chain fatty acids (C54 to C63)that are essential in cell wall formation of the human pathogen Mycobacterium tuberculosisas well as other mycobateria such as Mycobacterium leprae, and are associated with virulence[4]. InhA has been propsed as the target of the thioamide drugs, ethionamide (ETH) and isoniazid (INH), which have been used in treatment of mycobacterial infections [2].
Structure of InhAStructure of InhA
|
The inhA enzyme of M. tuberculosis is a homotetramer composed of a repeating subunit comprised of a single domain with a Rossmann Fold in the core that provides a NADH binding site[5]. The single domain can be broken down into two substructures that are connected by short peptide loop[1][5]. The overall structure exhibits α/β folding of a series of α strands flanking a central β sheet of multiple parallel β strands[5].
Substructure 1 of InhASubstructure 1 of InhA
consists of 6 parallel β strands and 4 α helices interwoven together to form a core α/β structure that contains the n-terminal domain[1]. The first substructure can be further broken down into two sections, the consisting of two β strands and two short α helicies [1]. The first section is connected to the by a β strand that crosses over the two domains, and leads into the second section initiating at the third α helix [1](A-3) is connected by a long loop to a 14 residue β strand that then leads into the fourth α helix [1]. A-4 then leads into a fifth strand β , followed by a 25 residue α helix , and into the final strand β [1].
Substructure 2 of InhASubstructure 2 of InhA
contains the c-terminal region of the molecule and consists of a small β strand , and two α helicies which are conected by a short five residue loop[1]. The C-terminal domain consits of two other α helicies [1].
InhA's Function in the Mycolic Acid PathwayInhA's Function in the Mycolic Acid Pathway

InhA plays a key role in the synthesis of fatty acids, particularly in M. tuberculosis which has type one fatty acid synthesis (FASI) and type two fatty acid synthesis (FASII) which together funtion in the synthesis of mycolic acids[7]. FASI synthesizes C16-18 and C24-26 fatty acids these are then sent to FASII promotes chain extention, forming long-chain meromycolic acids that are 56-64 carbons in length[8]. The final step in FASII is compleated by InhA which reduces 2-trans-enoyl-ACP's with chain lengths over twelve carbons in a NADP dependent manner where the hydride transfer precedes protonation[7][9].
The reaction takes place as follows. Intially NADH binds to the active site mediated by vander wall interactions with phenylalanine 41 (F41) and interations with lysine 165 [9][5]. Binding of NADH causes a conformational change in the Aspartate 42 and Arginine 43 side chains and an over all conformational change in InhA[5]. In addition tyrosine 158 (Y158) playes an important role in alinging the carbonyl substrate, in fact; rotaion about its Cα-Cβ bond by 60° brings it into a position where it can hydrogen bond to the carbonyl of the 2-trans enoyl-ACP and provide it with electrophilic stabalization[9]. Inha then reduces the 2-trans double bond of the substrate by forming a enoyl intermediate through the transfer of a hydride ion from NADH to the thrid carbon of the substrate, followed by protonation of the second carbon[5]. The biniding of both the substrate and the cofactor induces another conformational change in InhA that allows for the release of the meromycolic acid product[5]. The meromycolic acids undergo claisen condensation with a C26 fatty acid followed by reduction to a mature mycolic acid[8][5].
InhA and Thioamide DrugsInhA and Thioamide Drugs
Protein SuperfamillyProtein Superfamilly
InhA can also be classified into a family of short chain dehydrogenase/reductases (SDR). This family consists of proteins exhibiting a central core with a Rossmann fold that contains a NADH binding site. There are appoximatley 3000 primary strucures outlines in various sequence databases[10]. Examples of proteins in this family are listed below.
- 1bxk - DTDP-glucose 4,6-dehydratase -E. coli
- 1bsv - GDP-fructose synthetase in complex with NADPH - E. coli
- 1qrr - SQD1
- 1ae1 - Tyopinone reductase-I
- [[
- [[
- [[
- [[
- [[
- [[
InhA can be further claasified into the acyl carrier protein family. These proteins generally all function in the transport of substrates in a myrid of pathways, such as: the sysnthesis of polyketides and fatty acids[11]. Some examples of such proteins are listed below with links to their cooresponding proteopedia page.
- 3oic - FabL
- 1zhg - FabZ
- 3oig – BsENR I+NAD+INH
- 3oew - 2x22, 2x23, 1eny, 1enz – MtENR+NAD – Mycobacterium tuberculosis
- 2nsd – MtENR+NAD+piperidine derivative
- 1p44 - MtENR+NAD+indole derivative
- 1bvr - MtENR+NAD+fatty-acyl substrate
- 2ntv - ENR+PTH-NAD – Mycobacterium leprae
- 3oje – BcENR – Bacillus cereus
- 3ojf – BcENR+NADP+indole naphthyrididone
- 2foi – PfENR fragment+diaryl ether inhibitor
- 2qio - ENR+NAD+TCL – Bacillus anthracis
- 2p91 – ENR – Aquifex aeolicus
- 2pd3 – HpENR+TCL – Helicobacter pylori
- 1ve7 – ApAARE+p-nitrophenyl phosphate
- 1i2z - EcENR+NAD+imidazole derivative
Additional ResourcesAdditional Resources
ReferencesReferences
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Sacchettini, James (New Rochelle, NY) 1999 INHA crystals and three dimensional structure United States Albert Einstein College of Medicine of Yeshiva University (Bronx, NY) 5882878 http://www.freepatentsonline.com/5882878.html
- ↑ 2.0 2.1 2.2 Molle V, Gulten G, Vilcheze C, Veyron-Churlet R, Zanella-Cleon I, Sacchettini JC, Jacobs WR Jr, Kremer L. Phosphorylation of InhA inhibits mycolic acid biosynthesis and growth of Mycobacterium tuberculosis. Mol Microbiol. 2010 Dec;78(6):1591-605. doi:, 10.1111/j.1365-2958.2010.07446.x. Epub 2010 Nov 9. PMID:21143326 doi:10.1111/j.1365-2958.2010.07446.x
- ↑ Wang F, Langley R, Gulten G, Dover LG, Besra GS, Jacobs WR Jr, Sacchettini JC. Mechanism of thioamide drug action against tuberculosis and leprosy. J Exp Med. 2007 Jan 22;204(1):73-8. Epub 2007 Jan 16. PMID:17227913 doi:10.1084/jem.20062100
- ↑ Jmol - a paradigm shift in crystallographic visualization. J. Appl. Cryst. (2010). 43, 1250-1260 doi:https://dx.doi.org/10.1107/S0021889810030256
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 Dias MV, Vasconcelos IB, Prado AM, Fadel V, Basso LA, de Azevedo WF Jr, Santos DS. Crystallographic studies on the binding of isonicotinyl-NAD adduct to wild-type and isoniazid resistant 2-trans-enoyl-ACP (CoA) reductase from Mycobacterium tuberculosis. J Struct Biol. 2007 Sep;159(3):369-80. Epub 2007 May 3. PMID:17588773 doi:http://dx.doi.org/10.1016/j.jsb.2007.04.009
- ↑ Wilson M, DeRisi J, Kristensen HH, Imboden P, Rane S, Brown PO, Schoolnik GK. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc Natl Acad Sci U S A. 1999 Oct 26;96(22):12833-8. PMID:10536008
- ↑ 7.0 7.1 Gurvitz A, Hiltunen JK, Kastaniotis AJ. Function of heterologous Mycobacterium tuberculosis InhA, a type 2 fatty acid synthase enzyme involved in extending C20 fatty acids to C60-to-C90 mycolic acids, during de novo lipoic acid synthesis in Saccharomyces cerevisiae. Appl Environ Microbiol. 2008 Aug;74(16):5078-85. Epub 2008 Jun 13. PMID:18552191 doi:10.1128/AEM.00655-08
- ↑ 8.0 8.1 Bhatt A, Brown AK, Singh A, Minnikin DE, Besra GS. Loss of a mycobacterial gene encoding a reductase leads to an altered cell wall containing beta-oxo-mycolic acid analogs and accumulation of ketones. Chem Biol. 2008 Sep 22;15(9):930-9. PMID:18804030 doi:10.1016/j.chembiol.2008.07.007
- ↑ 9.0 9.1 9.2 Parikh S, Moynihan DP, Xiao G, Tonge PJ. Roles of tyrosine 158 and lysine 165 in the catalytic mechanism of InhA, the enoyl-ACP reductase from Mycobacterium tuberculosis. Biochemistry. 1999 Oct 12;38(41):13623-34. PMID:10521269
- ↑ Oppermann U, Filling C, Hult M, Shafqat N, Wu X, Lindh M, Shafqat J, Nordling E, Kallberg Y, Persson B, Jornvall H. Short-chain dehydrogenases/reductases (SDR): the 2002 update. Chem Biol Interact. 2003 Feb 1;143-144:247-53. PMID:12604210
- ↑ Rafi S, Novichenok P, Kolappan S, Zhang X, Stratton CF, Rawat R, Kisker C, Simmerling C, Tonge PJ. Structure of acyl carrier protein bound to FabI, the FASII enoyl reductase from Escherichia coli. J Biol Chem. 2006 Dec 22;281(51):39285-93. Epub 2006 Sep 29. PMID:17012233 doi:10.1074/jbc.M608758200