Sandbox Reserved 321: Difference between revisions
No edit summary |
No edit summary |
||
| Line 4: | Line 4: | ||
<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | <!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
'''InhA''' | |||
by Kelly Hrywkiw | by Kelly Hrywkiw | ||
| Line 19: | Line 19: | ||
==Structure== | ==Structure== | ||
The overall strucuture of the inhA enzyme of ''[http://en.wikipedia.org/wiki/Mycobacterium_tuberculosis Mycobacterium tuberculosis]'' consists of a single domain with two substructures<ref name ="making drugs for inhA">PMID:5882878</ref>. The first substructure can be further broken down into two sections consisting of two β strands (B-1 and B-2)and two short α helicies (A-1 and A-2)<ref name ="making drugs for inhA">PMID:5882878</ref>. | |||
==Role in the Mycolic Acid Pathway== | ==Role in the Mycolic Acid Pathway== | ||
Revision as of 03:13, 31 March 2011
| This Sandbox is Reserved from January 10, 2010, through April 10, 2011 for use in BCMB 307-Proteins course taught by Andrea Gorrell at the University of Northern British Columbia, Prince George, BC, Canada. |
To get started:
More help: Help:Editing |
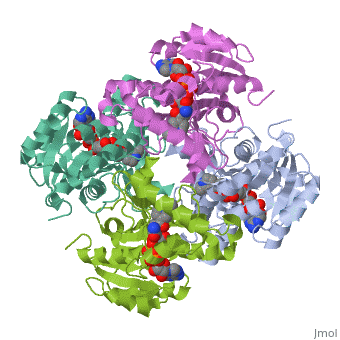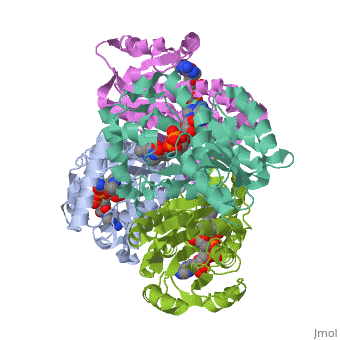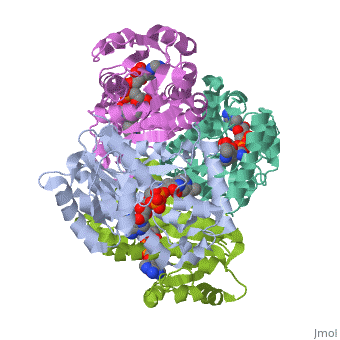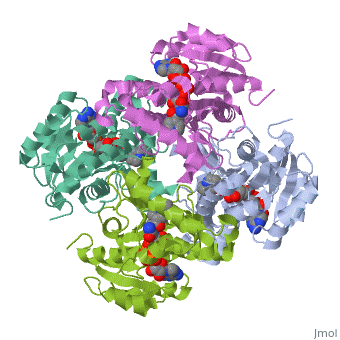
InhA
by Kelly Hrywkiw
| |||||||||
| 2h9i, resolution 2.20Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Gene: | inhA (Mycobacterium tuberculosis) | ||||||||
| Activity: | [acyl-carrier-protein_reductase_(NADH) Enoyl-[acyl-carrier-protein] reductase (NADH)], with EC number 1.3.1.9 | ||||||||
| Related: | 1zid | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
IntroductionIntroduction
The enzyme inhA is coded from the inhA gene that is simillar in sequence to the Salmonella typhimuriumgene which plays a role in fatty acid biosynthesis [1]. Inha is an NADH dependent trans enoyl-acyl ACP carrier protein that plays a role in the sysnthesis of Mycolic Acid, and is part of a short-chain dehydrogenase/reductase family [2][3]. Mycolic acids are long chain fatty acids that are essential in cell wall formation of the human pathogen Mycobacterium tuberculosisas well as other mycobateria such as Mycobacterium leprae[4]. Inha has been propsed as the target of the thionamide drugs, ethionamide (ETH) and isoniazid (INH), which have been used in treatment of mycobacterial infections [3].
StructureStructure
The overall strucuture of the inhA enzyme of Mycobacterium tuberculosis consists of a single domain with two substructures[1]. The first substructure can be further broken down into two sections consisting of two β strands (B-1 and B-2)and two short α helicies (A-1 and A-2)[1].
Role in the Mycolic Acid PathwayRole in the Mycolic Acid Pathway
Physiological FunctionPhysiological Function
Protein SuperfamillyProtein Superfamilly
ReferencesReferences
- ↑ 1.0 1.1 1.2 Strohmaier K, Streissle G, Clemm de Noronha S. [On the determination of size of early summer meningoencephalitis]. Arch Gesamte Virusforsch. 1965;17(2):300-3. PMID:5882878
- ↑ Wang F, Langley R, Gulten G, Dover LG, Besra GS, Jacobs WR Jr, Sacchettini JC. Mechanism of thioamide drug action against tuberculosis and leprosy. J Exp Med. 2007 Jan 22;204(1):73-8. Epub 2007 Jan 16. PMID:17227913 doi:10.1084/jem.20062100
- ↑ 3.0 3.1 Molle V, Gulten G, Vilcheze C, Veyron-Churlet R, Zanella-Cleon I, Sacchettini JC, Jacobs WR Jr, Kremer L. Phosphorylation of InhA inhibits mycolic acid biosynthesis and growth of Mycobacterium tuberculosis. Mol Microbiol. 2010 Dec;78(6):1591-605. doi:, 10.1111/j.1365-2958.2010.07446.x. Epub 2010 Nov 9. PMID:21143326 doi:10.1111/j.1365-2958.2010.07446.x
- ↑ Jmol - a paradigm shift in crystallographic visualization. J. Appl. Cryst. (2010). 43, 1250-1260 doi:https://dx.doi.org/10.1107/S0021889810030256