Sandbox423: Difference between revisions
No edit summary |
|||
| Line 46: | Line 46: | ||
Nick DeGraan-Weber, Jackie Dorhout, Rachael Jayne, Mike Reardon - flu neuraminidase | Nick DeGraan-Weber, Jackie Dorhout, Rachael Jayne, Mike Reardon - flu neuraminidase | ||
Brittany Forkus, Katie Geldart, Elizabeth Schutsky, Breanna Zerfas - undecided | |||
==Students looking for group members== | ==Students looking for group members== | ||
Revision as of 00:13, 31 March 2011
This sandbox is in use until August 1, 2011 for UMass Chemistry 423. Others please do not edit this page. Thanks!
Chem423 Team Projects: Understanding Drug Mechanisms
Project InstructionsProject Instructions
1. Team & Structure selection, due 4/1/11: All projects & names must be posted on the list below, linking to sandbox pages displaying initial structure.
- Form teams of 4 people; include both chemistry and chemical engineering majors on each team. Start by finding a protein-drug or nucleic acid-drug complex with a known structure in the pdb that interests your team. Check the list below to see if another team has already chosen this complex. If not, start a new sandbox page (just try sandbox## in the search box to find an unused number) and add a link for your team/protein to our class list below (use editing button above (Ab) or follow my model).
- Copy the message at the top of this page into your sandbox page to "reserve" your sandbox for this course.
- Find the pdb id for your protein-drug complex in the Protein Data Bank. In your sandbox page click"edit this page" (top) and follow the directions to insert your rotating structure on your page.
2. Project completion, due 4/22/11 Your proteopedia page should be organized into the following 5 required sections, with each team member responsible for one of these sections of the team project.
a. Introduction
- Introduce the protein function and the disease treated by the drug. This must be written in your own words with citations to your sources.You cannot include a copyrighted figure unless you request permission to use it.
b. Overall structure
- Describe the overall structure of your protein in words and make "green scenes" to illustrate your points. What elements of secondary structure are present (ie 5 alpha helices and 2 beta strands) and how are they organized? Below I illustrate the start of an "overall structure" section on GFP. Additional description and green scenes could illustrate the polar/nonpolar distrubution of amino acids (is the inside of the barrel polar or nonpolar?), packing of amphipathic elements, etc.
c. Drug binding site
- Describe features of the drug binding site in words and make "green scenes" to illustrate your points. Show the interactions that stabilize binding of this molecule to the protein (ie H bonds).
d. Additional features
- Describe and use green scenes to illustrate additional features of the protein. What you do here depends on what information is available. If a structure of the protein-substrate complex is available, you could compare protein interactions with the substrate vs. with the drug. If the drug is a transition state inhibitor, explain and illustrate that (eg include a reaction scheme with structures of the substrate, transition state and product).
e. Credits -- at the end list who did which portion of the project:
- Introduction -- name of team member
- Overall structure -- name of team member
- Drug binding site -- name of team member
- Additional features -- name of team member
3. In-class presentations, to be announced
ExampleExample
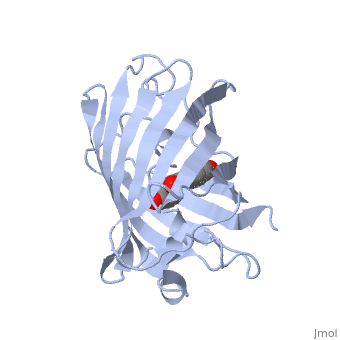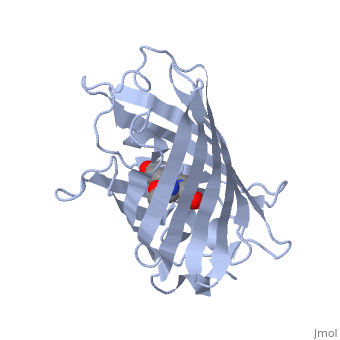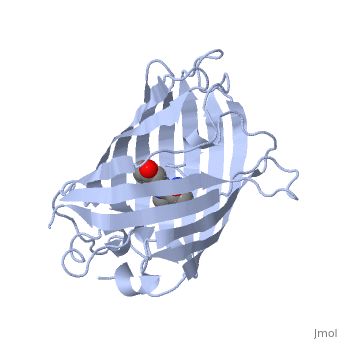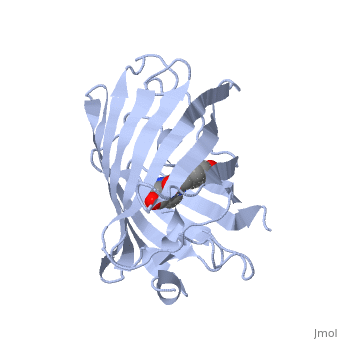
One green scene for "Overall Structure" section:
| |||||||||
| 1ema, resolution 1.90Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Non-Standard Residues: | , | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
The of green fluorescent protein is an 11-stranded antiparallel beta barrel (yellow) surrounding a central helix (pink). The chromophore is part of the central helix. Information from 1ema.
Project Teams, Topics, and LinksProject Teams, Topics, and Links
sample entry: student1, student2, student3, student4 - HIV protease sandbox1
Nick DeGraan-Weber, Jackie Dorhout, Rachael Jayne, Mike Reardon - flu neuraminidase
Brittany Forkus, Katie Geldart, Elizabeth Schutsky, Breanna Zerfas - undecided
Students looking for group membersStudents looking for group members
Each group should contain at least one person from a different primary major (typically Chemistry or Chemical Engineering) than the rest.
List yourself + your major, list partial groups looking for members, list your complex if you have chosen one. Contact others to form a group.
Jaclyn Somadelis, Chemical Engineering
Justin Srodulski, Chemical Engineering
Max Moulton, Sally Stras, Jordan Schleeweis, all chemical engineering, need chem major -- HIV reverse transcriptase
Lucia Tringali and Shaina Boyle - Both Chem majors in need of Chemical engineering partner(s).
Help EditingHelp Editing
Hint: Ctl-click or right-click on links below and select "Open Link in New Window"
- You can use the edit button on any page to find out how other users created effects that you see.
[General help with Wiki editing], plus more [Wiki Text examples]
Some of the above are for help editing Wikipedia pages, but the syntax is mostly the same. Proteopedia ADDS protein stuff to the WikiMedia markup language, which powers both WEB sites.