Ramachandran Plot: Difference between revisions
No edit summary |
No edit summary |
||
| Line 49: | Line 49: | ||
#In the Jmol applet showing the 3D structure on the page, click on the Jmol logo (or frank) in the bottom right corner. | #In the Jmol applet showing the 3D structure on the page, click on the Jmol logo (or frank) in the bottom right corner. | ||
#When the menu comes up, select '''<code>Console</code>''' | #When the menu comes up, select '''<code>Console</code>''' | ||
#Click in the lower text window of the console that comes up and type the command '''<code>Ramachandran</code>''', followed by the return key. | #Click in the lower text window of the console that comes up and type the command '''<code>plot Ramachandran</code>''', followed by the return key. | ||
#After some processing the Ramachandran plot will be visible and you can hover over and click on the points in the plot just as you can with atoms in a Jmol scene window. (To return to the model, an easy solution is to reload the page or open a new browser instance of that page.) | #After some processing the Ramachandran plot will be visible and you can hover over and click on the points in the plot just as you can with atoms in a Jmol scene window. (To return to the model, an easy solution is to reload the page or open a new browser instance of that page.) | ||
Revision as of 20:19, 9 October 2010
|
A Ramachandran plot is a plot of the torsional angles - psi (ψ)and phi (φ) - of the residues (amino acids) contained in a peptide. The plot was developed in 1963 by G. N. Ramachandran, et. al.[1] by plotting the ψ values on the y-axis and the φ values on the x-axis. Plotting the torsional angles in this way graphically shows which combination of angles are possible. The torsional angles of each residue in a peptide define the geometry of its attachment to its two adjacent residues by positioning its planar peptide bond relative to the two adjacent planar peptide bonds, thereby the torsional angles determine the conformation of the residues and the peptide. Many of the angle combinations, and therefore the conformations of residues, are not possible because of steric hindrance. By making a Ramachandran plot protein structural scientist can determine which torsional angles are permitted and can obtain insight into the structure of peptides. The scene on the right is the Ramachandran plot of ribonuclease H.
Secondary structure plot regionsSecondary structure plot regions
Secondary structures of a peptide are segments of the peptide that have ordered and repetitive structure, and the repetitive structure is due to a repetitive conformation of the residues and, ultimately, repetitive values of ψ and φ. The different secondary structures can be distinguished by their range of ψ and φ values with the values of different secondary structures mappng to different regions of the Ramachandran plot. Two common examples of secondary structure are illustrated below.
α-helixα-helix
| |||||||
The scene on the right shows the axis of the α-helix rotating in the y-plane. When viewing the helix on end, observe the open center of the helix. are drawn on some of the peptide bonds to emphasize that in an α-helix the planar peptide bonds rotate about the axis of the helix. The of this peptide has points clustered about the values of ψ= -47o and φ= -57o which are the average values for α-helices. of two other helical segments demonstrates that data from all three appear in one large cluster and that the helical segments can not be distinguished by the differences in their ψ and φ values.
β-sheetsβ-sheets
a two segment twisted β-sheets. of the peptide bonds. Most β-sheets in globular proteins are twisted sheets which do not have even parallel pleads. of β-sheets. The of this twisted sheet has points clustered about the values of ψ= -135o and φ= +135o which are the average values for twisted sheets. of three other sheet segments more clearly defines the area in which values for twisted sheets are located.
Plot regions limited by steric hindrancePlot regions limited by steric hindrance
| |||||||
Most combinations of ψ and φ are sterically forbidden, as illustrated in the tripeptide, Glu-Ser-Ala. With Ser having values of ψ = -116o and φ = 55o the Ser side chain is in contact with the Ala, colored blue. In plots of native peptides the data points will form clusters in the several areas in which steric hindrance does not occur. These regions are illustrated in the Figure on the left. The core regions (blue in the Figure) contain the most favorable combinations of ψ and φ and contain the greatest number of points.
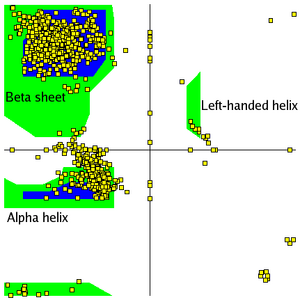
Plots of some proteins contain a small third core region in the upper right quadrant. The allowed regions (green in the Figure) can be located around the core regions or can be unassociated with a core region, but they contain fewer data points than the core regions. The generous regions (not shown in the Figure)extend beyond the allowed regions. The remaining areas are the disallowed regions. Observe that the data point -116o, 55o for Ser of the above tripeptide would fall in the lower right quadrant which contains only a disallowed region. If the Ser in the tripeptide has ψ and φ values of -47o and -57o which are in the core region in the lower left, the is rotated away from the Ala and is not in contact with the Ala. Most, if not all, of the points in the above plots for α-helix and β-sheet are in one of the core areas.
GlycineGlycine
Since Gly has only a hydrogen as a side chain, steric hindrance is not as likely to occur as ψ and φ are rotated through a series of values. The with Gly having ψ and φ values of -116o and +55o respectively, does not show the steric hindrance that the Glu-Ser-Ala had. For that reason Gly will frequently plot in the disallowed region of a Ramachandran plot. Nearly all of the data points in the disallowed region in the above Figure are Gly points.
Functionally relevant residuesFunctionally relevant residues
Functionally relevant residues may occasionally have torsion angles that plot to the disallowed regions of a Ramachandran plot. The specific geometry of these functionally relevant residues, while energetically unfavorable, may be important for the protein's function, catalytic or otherwise.
Plots of proteinsPlots of proteins
|
: Secondary structure consist of α-helix, loops and ordered, nonrepetitive structures.
: Red data points outside of the area expected for α-helix most likely involve residues at the end of the α-helix because often these have angle values that are not typical for α-helix. White points are those for loops and ordered, nonrepetitive structures. The few residues that map to the disallowed region are Gly.
after viewing plot
: Twisted β-sheet with small segments of α-helix.
: Most of the yellow points are located in the area for twisted β-sheets where one would expect them, and again the points in the disallowed region are Gly.
after viewing plot.
: Close to equal amounts of α-helix, β-sheet, and ordered, nonrepetitive structures. One important exception to Gly in the disallowed region is Ser:200. Locate this residue that is located in a disallowed region (lower right quadrant). An interesting aspect concerning Ser:200 is that it is one of a triad of residues that are part of the catalytic site and are involved in the catalytic action of this enzyme. The unique φ and ψ values for Ser:200 is the major factor in positioning the side chain so that it can participate in the catalysis.
after viewing plot.
OTHER ENTRIES IN PROTEOPEDIA
Interactive Ramachandran plots can be generated for any entry in Proteopedia with the use of a typed Jmol command[2]:
- For example, in a new browser window open the entry in Proteopedia for 1bhl
- In the Jmol applet showing the 3D structure on the page, click on the Jmol logo (or frank) in the bottom right corner.
- When the menu comes up, select
Console - Click in the lower text window of the console that comes up and type the command
plot Ramachandran, followed by the return key. - After some processing the Ramachandran plot will be visible and you can hover over and click on the points in the plot just as you can with atoms in a Jmol scene window. (To return to the model, an easy solution is to reload the page or open a new browser instance of that page.)
This method to generate interactive Ramachandran plots will also work for other instances of the Jmol applet elsewhere on the web as long as the version of the Jmol is 11.4 or greater.
If you just need to report φ and ψ values for a few residues, use the Scene Authoring Tools to select the residues of interest and enter the command draw RAMACHANDRAN in the console.
NotesNotes
- ↑ RAMACHANDRAN GN, RAMAKRISHNAN C, SASISEKHARAN V (July 1963). "Stereochemistry of polypeptide chain configurations". J. Mol. Biol. 7: 95–9. PMID 13990617
- ↑ Command defined at site for official Jmol documentation
External LinksExternal Links
Ramachandran Plot at Wikipedia
α-helix at Wikipedia
β-sheets at Wikipedia
Another example of a Ramachandran Plot showing the different regions. at the European Bioinformatics Institute (EBI)