Nitrogenase: Difference between revisions
No edit summary |
No edit summary |
||
| Line 17: | Line 17: | ||
At the bottom of the protein, where the Fe protein comes into contact with the FeMo protein, is a <scene name='Sandbox_10/1n2c_fes_cluster_cys/1'>4Fe:4S cluster</scene>, held in place by cysteines. This cluster accepts electrons from ferredoxin and gives electrons to the FeMo protein. | At the bottom of the protein, where the Fe protein comes into contact with the FeMo protein, is a <scene name='Sandbox_10/1n2c_fes_cluster_cys/1'>4Fe:4S cluster</scene>, held in place by cysteines. This cluster accepts electrons from ferredoxin and gives electrons to the FeMo protein. | ||
When the Fe protein is bound to the FeMo protein (<scene name='Sandbox_10/1n2c_single_complex/2'>zoom out</scene>), ATP is hydrolyzed and electrons that were transferred to the 4Fe:4S cluster of the Fe protein by ferredoxin are transferred to the FeMo protein. The crystal structure of the complete nitrogenase complex reveals how the binding of the Fe and FeMo proteins, the hydrolysis of ATP, and the transfer of electrons are all coupled. | When the Fe protein is bound to the FeMo protein (<scene name='Sandbox_10/1n2c_single_complex/2'>zoom out</scene>), ATP is hydrolyzed and electrons that were transferred to the 4Fe:4S cluster of the Fe protein by ferredoxin are transferred to the FeMo protein. The crystal structure of the complete nitrogenase complex reveals how the binding of the Fe and FeMo proteins, the hydrolysis of ATP, and the transfer of electrons are all coupled. The figure below summarizes how these processes are linked. | ||
[[Image:clip_image002.jpg]] | |||
the two subunits of the Fe protein are pushed closer together. When this happens | the two subunits of the Fe protein are pushed closer together. When this happens | ||
Revision as of 01:24, 21 June 2010
Please do NOT make changes to this sandbox. Sandboxes 10-30 are currently reserved by Prof. Sheila Jaswal at Amherst College.
Nitrogenase
|
Nitrogenase is an enzyme that fixes atmospheric nitrogen (N2) into ammonia. Though abundantly present in the atmosphere, most organisms cannot utilize N2 directly, and must instead take it in through other forms, like ammonia or nitrate. The triple bond in N2 is highly resistant to changes in oxidation state, and nitrogenases, found only in nitrogen-fixing bacteria, are the only proteins capable of reducing N2 to ammonia.
Nitrogenase catalyzes the following reaction:
N2 + 8 H+ + 16 MgATP + 8 e- → 2NH3 + H2 + 16 MgADP + 16 Pi
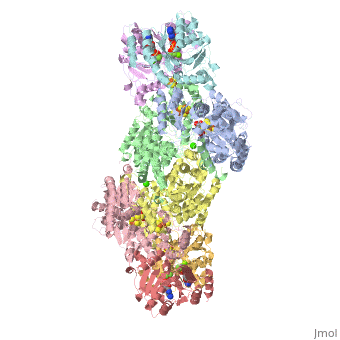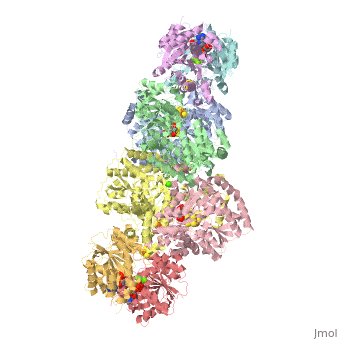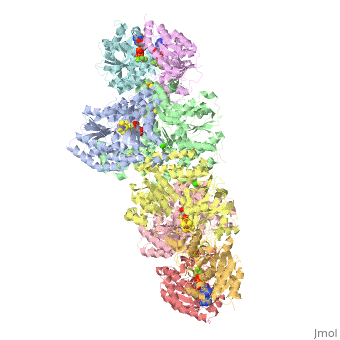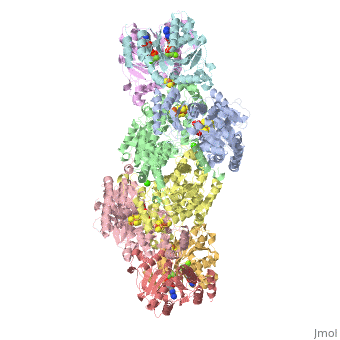
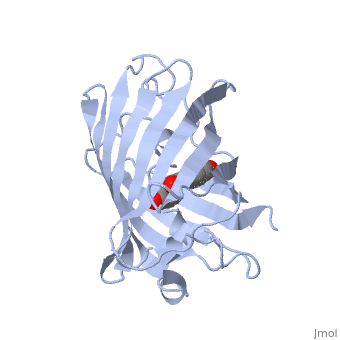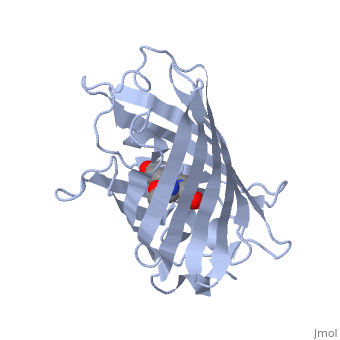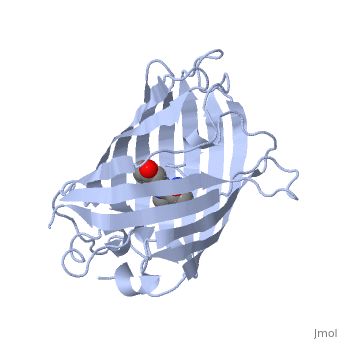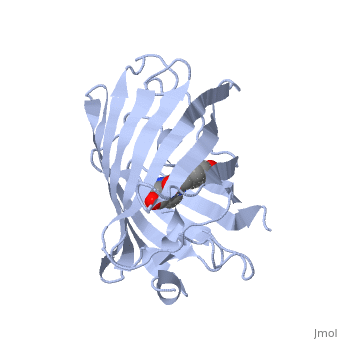
Two different proteins comprise the nitrogenase complex. The FeMo protein binds substrate and reduces H+ and N2 to H2 and ammonia, while the Fe protein receives electrons from ferredoxin, hydrolyzes ATP, and reduces the FeMo protein. To the right is shown a crystal structure (PDB entry 1n2c) where two complexes of FeMo protein bound to Fe protein were crystallized together. Click here to see only .
The is here bound to two , an analog for the planar transition state of ATP hydrolysis. The motif that binds ATP is a conserved nucleotide binding motif called Walker's motif A. Coloring by , the nucleotide binding pocket is clear.
At the bottom of the protein, where the Fe protein comes into contact with the FeMo protein, is a , held in place by cysteines. This cluster accepts electrons from ferredoxin and gives electrons to the FeMo protein.
When the Fe protein is bound to the FeMo protein (), ATP is hydrolyzed and electrons that were transferred to the 4Fe:4S cluster of the Fe protein by ferredoxin are transferred to the FeMo protein. The crystal structure of the complete nitrogenase complex reveals how the binding of the Fe and FeMo proteins, the hydrolysis of ATP, and the transfer of electrons are all coupled. The figure below summarizes how these processes are linked.
the two subunits of the Fe protein are pushed closer together. When this happens
Green fluorescent protein (GFP), originally isolated from the jellyfish Aequorea victoria (PDB entry 1ema), fluorsceses green (509nm) when exposed to blue light (395nm and 475nm). It is one of the most important proteins used in biological research because it can be used to tag otherwise invisible gene products of interest and thus observe their existence, location and movement.
Exploring the StructureExploring the Structure
|
GFP is a beta barrel protein with 11 beta sheets. It is a 26.9kDa protein made up of 238 amino acids. The , responsible for the fluorescent properties of the protein, is buried inside the beta barrel as part of the central alpha helix passing through the barrel. The chromophore forms via spontaneous cyclization and oxidation of three residues in the central alpha helix: -Thr65 (or Ser65)-Tyr66-Gly67. This cyclization and oxidation creates the chromophore's five-membered ring via a new bond between the threonine and the glycine residues.[1]