Jake Ezell Sandbox 2: Difference between revisions
Jake Ezell (talk | contribs) |
Jake Ezell (talk | contribs) |
||
| Line 1: | Line 1: | ||
==Malate Dehydrogenase== | ==Malate Dehydrogenase== | ||
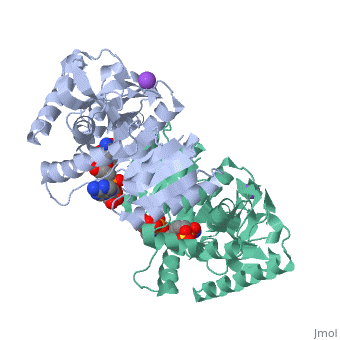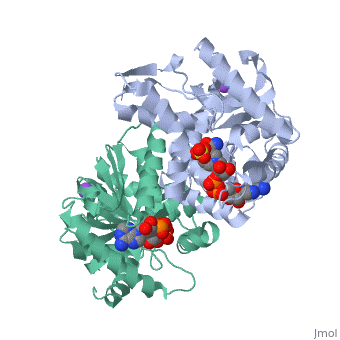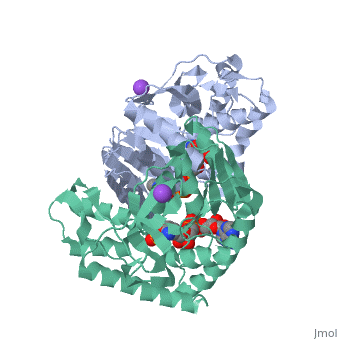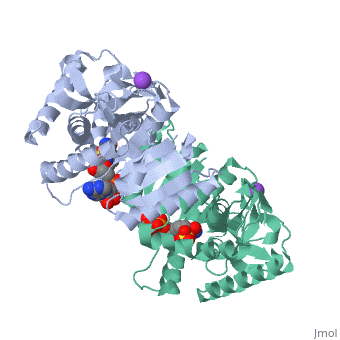
Malate Dehydrogenase (MDH)(PDB entry [http://www.pdb.org/pdb/explore/explore.do?structureId=2X0I 2x0i]) is most known for its role in the metabolic pathway in the tricarboxylic acid cycle[http://en.wikipedia.org/wiki/Citric_acid_cycle]; however, the enzyme is also in many other metabolic pathways such as glyoxylate bypass, amino acid synthesis, glucogenesis, and oxidation/reduction balance <ref>PMID:12537350</ref> . It is classified as a oxidoreductase[http://en.wikipedia.org/wiki/Oxidoreductase]. Malate Dehydrogenase has been extensively studied due to its many isozymes <ref>PMID:20173310</ref>. The enzyme exists in two places inside a cell, in the mitochondria and cytoplasm. In the mitochondria, the enzyme catalyzes the reaction of malate to oxaloacetate; but in the cytoplasm, the enzyme catalyzes oxaloacetate to malate to allow transport <ref>PMID:20173310</ref> . This conversion is an essential catalytic step in each different metabolic mechanism. The enzyme malate dehydrogenase is composed of either a dimer or tetramer <ref>PMID: 9834842<ref/> . During catalysis, the enzyme subunits are non-cooperative between active sites. The mitochondrial MDH is allosterically controlled by citrate, but no other known metabolic regulation mechanisms have been discovered. | Malate Dehydrogenase (MDH)(PDB entry [http://www.pdb.org/pdb/explore/explore.do?structureId=2X0I 2x0i]) is most known for its role in the metabolic pathway in the tricarboxylic acid cycle[http://en.wikipedia.org/wiki/Citric_acid_cycle]; however, the enzyme is also in many other metabolic pathways such as glyoxylate bypass, amino acid synthesis, glucogenesis, and oxidation/reduction balance <ref>PMID:12537350</ref>. It is classified as a oxidoreductase[http://en.wikipedia.org/wiki/Oxidoreductase]. Malate Dehydrogenase has been extensively studied due to its many isozymes <ref>PMID:20173310</ref>. The enzyme exists in two places inside a cell, in the mitochondria and cytoplasm. In the mitochondria, the enzyme catalyzes the reaction of malate to oxaloacetate; but in the cytoplasm, the enzyme catalyzes oxaloacetate to malate to allow transport <ref>PMID:20173310</ref>. This conversion is an essential catalytic step in each different metabolic mechanism. The enzyme malate dehydrogenase is composed of either a dimer or tetramer <ref>PMID:9834842<ref/>. During catalysis, the enzyme subunits are non-cooperative between active sites. The mitochondrial MDH is allosterically controlled by citrate, but no other known metabolic regulation mechanisms have been discovered. | ||
The secondary structure of a single subunit contains a <scene name='Malate_dehydrogenase/Beta_sheeting_backbone/1'>9 beta sheet parallel backbone</scene> wrapped by <scene name='Malate_dehydrogenase/Alpha_wrapping_betas/1'>9 large alpha helices</scene>. Near the sodium bound end, 4 small anti-parallel beta sheets and 1 small alpha helix enable a turn in the residue chain<scene name='Malate_dehydrogenase/Small_turn/1'>(small turn)</scene>. Opposite the sodium bound ligand, 6 alpha helices point towards a common point, three on each side of the beta sheet backbone. The alpha helices form a <scene name='Jake_Ezell_Sandbox_2/Small_groove_nad/1'>small groove</scene> for a NAD+ cofactor to attach near the beta sheeting. The structure most nearly resembles an alternating alpha/beta classification. As for the 3D structure, the enzyme forms a sort of | The secondary structure of a single subunit contains a <scene name='Malate_dehydrogenase/Beta_sheeting_backbone/1'>9 beta sheet parallel backbone</scene> wrapped by <scene name='Malate_dehydrogenase/Alpha_wrapping_betas/1'>9 large alpha helices</scene>. Near the sodium bound end, 4 small anti-parallel beta sheets and 1 small alpha helix enable a turn in the residue chain<scene name='Malate_dehydrogenase/Small_turn/1'>(small turn)</scene>. Opposite the sodium bound ligand, 6 alpha helices point towards a common point, three on each side of the beta sheet backbone. The alpha helices form a <scene name='Jake_Ezell_Sandbox_2/Small_groove_nad/1'>small groove</scene> for a NAD+ cofactor to attach near the beta sheeting. The structure most nearly resembles an alternating alpha/beta classification. As for the 3D structure, the enzyme forms a sort of | ||
Revision as of 10:56, 22 March 2010
Malate DehydrogenaseMalate Dehydrogenase
Malate Dehydrogenase (MDH)(PDB entry 2x0i) is most known for its role in the metabolic pathway in the tricarboxylic acid cycle[1]; however, the enzyme is also in many other metabolic pathways such as glyoxylate bypass, amino acid synthesis, glucogenesis, and oxidation/reduction balance [1]. It is classified as a oxidoreductase[2]. Malate Dehydrogenase has been extensively studied due to its many isozymes [2]. The enzyme exists in two places inside a cell, in the mitochondria and cytoplasm. In the mitochondria, the enzyme catalyzes the reaction of malate to oxaloacetate; but in the cytoplasm, the enzyme catalyzes oxaloacetate to malate to allow transport [3]. This conversion is an essential catalytic step in each different metabolic mechanism. The enzyme malate dehydrogenase is composed of either a dimer or tetramer Cite error: Closing </ref> missing for <ref> tag.
The evolutionary past of MDH shows a divergence to form lactate dehydrogenase (LDH) which functions in a very similar way to MDH. Although there is a very low sequence conservation among MDH and LDH’s [3] the structure of the enzyme has remained relatively conserved. The key difference between the two is in the substrate: LDH catalyzes pyruvate to lactate.
| |||||||
| 2x0i, resolution 2.91Å () | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||
| Activity: | Malate dehydrogenase, with EC number 1.1.1.37 | ||||||
| Related: | 2x06, 2x0j, 2x0n, 2x0r, 2x0s | ||||||
| |||||||
| |||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
ReferencesReferences
- ↑ Minarik P, Tomaskova N, Kollarova M, Antalik M. Malate dehydrogenases--structure and function. Gen Physiol Biophys. 2002 Sep;21(3):257-65. PMID:12537350
- ↑ Matsuda T, Takahashi-Yanaga F, Yoshihara T, Maenaka K, Watanabe Y, Miwa Y, Morimoto S, Kubohara Y, Hirata M, Sasaguri T. Dictyostelium Differentiation-Inducing Factor-1 Binds to Mitochondrial Malate Dehydrogenase and Inhibits Its Activity. J Pharmacol Sci. 2010 Feb 20. PMID:20173310
- ↑ Matsuda T, Takahashi-Yanaga F, Yoshihara T, Maenaka K, Watanabe Y, Miwa Y, Morimoto S, Kubohara Y, Hirata M, Sasaguri T. Dictyostelium Differentiation-Inducing Factor-1 Binds to Mitochondrial Malate Dehydrogenase and Inhibits Its Activity. J Pharmacol Sci. 2010 Feb 20. PMID:20173310