Succinyl-CoA synthetase: Difference between revisions
Lucas Hamlow (talk | contribs) No edit summary |
Lucas Hamlow (talk | contribs) No edit summary |
||
| Line 5: | Line 5: | ||
==Structure== | ==Structure== | ||
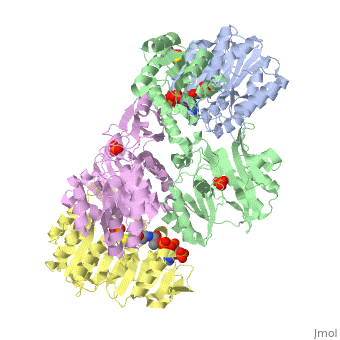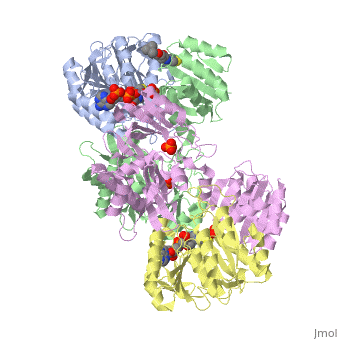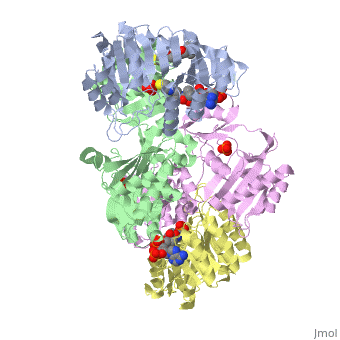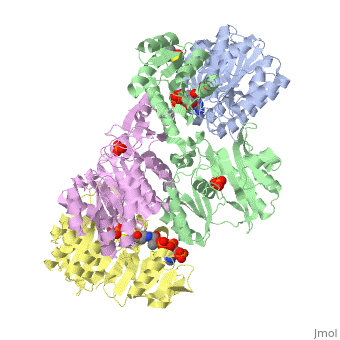
'''Succinyl-CoA synthetase''' is a tetramer with an active site on each subunit. This can be seen in the Jmol representation (the PDB code is 1CQJ). The two subunits are denoted alpha and beta. A phosphorylated histidine intermediate <scene name='Lucas_Hamlow_sandbox_1/Active_histidine_residue/1'>(HIS 246 alpha)</scene> is responsible for the dephosphorylation of ATP and it is suspected that there is another active site on the beta subunit that is responsible for the continued catalysis of the reaction. There is a suspected nucleotide binding site on the N-terminal of the beta subunit which would imply that there are two active sites roughly 35 A apart from each other and that the HIS 246 alpha loop moves between them during catalysis.<ref>PMID:10353839</ref> | '''Succinyl-CoA synthetase''' is a tetramer with an active site on each subunit. This can be seen in the Jmol representation (the PDB code is 1CQJ). The two subunits are denoted alpha and beta. A phosphorylated histidine intermediate <scene name='Lucas_Hamlow_sandbox_1/Active_histidine_residue/1'>(HIS 246 alpha)</scene> is responsible for the dephosphorylation of ATP and it is suspected that there is another active site on the beta subunit that is responsible for the continued catalysis of the reaction. There is a suspected nucleotide binding site on the N-terminal of the beta subunit which would imply that there are two active sites roughly 35 A apart from each other and that the HIS 246 alpha loop moves between them during catalysis.<ref>PMID:10353839</ref> | ||
On the alpha subunit it has been shown that <scene name='Lucas_Hamlow_sandbox_1/Active_his_with_glu_208/1'>GLU 208 alpha</scene> interacts with the active HIS 246 alpha residue in the phosphorylated and dephosphorylated enzyme and it is supposed that GLU 197 beta serves a similar purpose on the beta subunit.<ref>PMID:11781092</ref> | On the alpha subunit it has been shown that <scene name='Lucas_Hamlow_sandbox_1/Active_his_with_glu_208/1'>GLU 208 alpha</scene> interacts in some way with the active HIS 246 alpha residue in the phosphorylated and dephosphorylated enzyme and it is supposed that GLU 197 beta serves a similar purpose on the beta subunit.<ref>PMID:11781092</ref> | ||
==Mechanism== | ==Mechanism== | ||
A cooperative binding catalysis mechanism has been proposed and it has been shown that binding of ATP at one catalytic site promotes catalytic activity at another catalysis site.<ref>PMID:6997289</ref> It has been shown that the enzyme will bind with ATP in the presence of Mg+2 to form a complex containing 2 ATP residues as well as 2 phosphoric acid residues, after incubation this the complex converts to another one containing 4 phosphoric residues per protein. Only the second complex reacts with succinate and CoA to form the succinyl-CoA complex which then releases as many phosphoric residues as bound succinate.<ref>PMID:570066</ref> | A cooperative binding catalysis mechanism has been proposed and it has been shown that binding of ATP at one catalytic site promotes catalytic activity at another catalysis site.<ref>PMID:6997289</ref> It has been shown that the enzyme will bind with ATP in the presence of Mg+2 to form a complex containing 2 ATP residues as well as 2 phosphoric acid residues, after incubation this the complex converts to another one containing 4 phosphoric residues per protein. Only the second complex reacts with succinate and CoA to form the succinyl-CoA complex which then releases as many phosphoric residues as bound succinate.<ref>PMID:570066</ref> A transfer of the phosphoric residue from the first active site is seen to be coordinate with a transfer of a phosphoric residue to the second active site suggesting again a cooperative binding catalysis. This cooperative catalysis means that the presence of ATP or ADP can be both activating and inhibiting depending on the stage of catalysis they interact with the enzyme. | ||
==Bound Form of Succinyl-CoA synthetase== | ==Bound Form of Succinyl-CoA synthetase== | ||
Revision as of 07:53, 22 March 2010
|
Succinyl-Coa synthetase catalyzes the reversible reaction of succinyl-CoA + NDP + Pi <-> succinate + CoA + NTP (where N is either adenosine or guanosine. It can be found in Escherichia coli.
StructureStructure
Succinyl-CoA synthetase is a tetramer with an active site on each subunit. This can be seen in the Jmol representation (the PDB code is 1CQJ). The two subunits are denoted alpha and beta. A phosphorylated histidine intermediate is responsible for the dephosphorylation of ATP and it is suspected that there is another active site on the beta subunit that is responsible for the continued catalysis of the reaction. There is a suspected nucleotide binding site on the N-terminal of the beta subunit which would imply that there are two active sites roughly 35 A apart from each other and that the HIS 246 alpha loop moves between them during catalysis.[1] On the alpha subunit it has been shown that interacts in some way with the active HIS 246 alpha residue in the phosphorylated and dephosphorylated enzyme and it is supposed that GLU 197 beta serves a similar purpose on the beta subunit.[2]
MechanismMechanism
A cooperative binding catalysis mechanism has been proposed and it has been shown that binding of ATP at one catalytic site promotes catalytic activity at another catalysis site.[3] It has been shown that the enzyme will bind with ATP in the presence of Mg+2 to form a complex containing 2 ATP residues as well as 2 phosphoric acid residues, after incubation this the complex converts to another one containing 4 phosphoric residues per protein. Only the second complex reacts with succinate and CoA to form the succinyl-CoA complex which then releases as many phosphoric residues as bound succinate.[4] A transfer of the phosphoric residue from the first active site is seen to be coordinate with a transfer of a phosphoric residue to the second active site suggesting again a cooperative binding catalysis. This cooperative catalysis means that the presence of ATP or ADP can be both activating and inhibiting depending on the stage of catalysis they interact with the enzyme.
Bound Form of Succinyl-CoA synthetaseBound Form of Succinyl-CoA synthetase
|
The form of succinyl-CoA synthetase shown here (PDB code 1CQI) is the bound form with two (the ADP is highlighted and the Mg is in green).[5] As seen here, the residues on both the alpha and beta subunits are present and interacting with the lone Pi group while the ADP group is bound elsewhere on the subunit.
- ↑ Joyce MA, Fraser ME, Brownie ER, James MN, Bridger WA, Wolodko WT. Probing the nucleotide-binding site of Escherichia coli succinyl-CoA synthetase. Biochemistry. 1999 Jun 1;38(22):7273-83. PMID:10353839 doi:10.1021/bi990527s
- ↑ Fraser ME, Joyce MA, Ryan DG, Wolodko WT. Two glutamate residues, Glu 208 alpha and Glu 197 beta, are crucial for phosphorylation and dephosphorylation of the active-site histidine residue in succinyl-CoA synthetase. Biochemistry. 2002 Jan 15;41(2):537-46. PMID:11781092
- ↑ Bild GS, Janson CA, Boyer PD. Subunit interaction during catalysis. ATP modulation of catalytic steps in the succinyl-CoA synthetase reaction. J Biol Chem. 1980 Sep 10;255(17):8109-15. PMID:6997289
- ↑ Mikeladze DG, Matveeva LN, Severin SE. [Reaction mechanism of succinyl CoA synthetase from pigeon thoracic muscle] Biokhimiia. 1978 Aug;43(8):1458-67. PMID:570066
- ↑ Joyce MA, Fraser ME, James MN, Bridger WA, Wolodko WT. ADP-binding site of Escherichia coli succinyl-CoA synthetase revealed by x-ray crystallography. Biochemistry. 2000 Jan 11;39(1):17-25. PMID:10625475