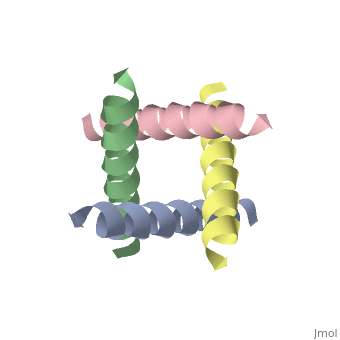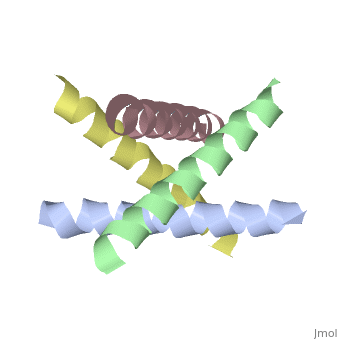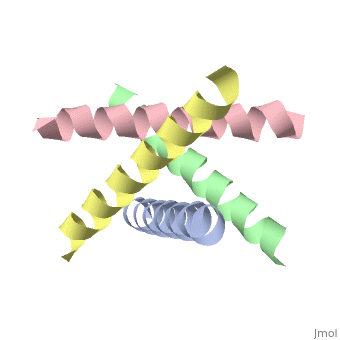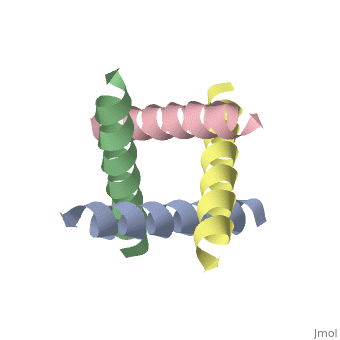M2 Proton Channel: Difference between revisions
Sarah Henke (talk | contribs) No edit summary |
Sarah Henke (talk | contribs) No edit summary |
||
| Line 3: | Line 3: | ||
== Background == | == Background == | ||
The M2 proton channel is a key protein that leads to viral infection [Takeuchi et al]. The M2 proton channel acidifies the viron which allows the viral matrix protein (M1) to disassociate from the ribonucleoprotein (RNP) [wu et al]. This allows the RNP to be transported to the nucleus of the cell [wu et al]. Several recent studies have looked at the effects of <scene name='User:Sarah_Henke/Sandbox_1/Amantadine/1'>amantadine</scene> <ref>PMID:18235504</ref> and <scene name='User:Sarah_Henke/Sandbox_1/Rimantadine/1'>rimantadine</scene> [Schnell et al] on inhibiting the transfer of protons through the M2 channel <ref name="Stouffer" />. It has been found that M2 is resistant to these two drugs in 90% of humans, birds and pigs <ref name="Stouffer" />. Understanding the structure and function of this proton channel is necessary in solving the resistance problem <ref name="Stouffer" />. | The M2 proton channel is a key protein that leads to viral infection [Takeuchi et al]. The M2 proton channel acidifies the viron which allows the viral matrix protein (M1) to disassociate from the ribonucleoprotein (RNP) [wu et al]. This allows the RNP to be transported to the nucleus of the cell [wu et al]. Several recent studies have looked at the effects of <scene name='User:Sarah_Henke/Sandbox_1/Amantadine/1'>amantadine</scene> <ref>PMID:18235504</ref> and <scene name='User:Sarah_Henke/Sandbox_1/Rimantadine/1'>rimantadine</scene> [Schnell et al] on inhibiting the transfer of protons through the M2 channel <ref name="Stouffer" />. It has been found that M2 is resistant to these two drugs in 90% of humans, birds and pigs <ref name="Stouffer" />. Understanding the structure and function of this proton channel is necessary in solving the resistance problem <ref name="Stouffer AL" />. | ||
== Structure == | == Structure == | ||
The M2 proton channel from influenza A is 97 amino acid residues and forms a 24-residue N-terminal extracellular domain, a 19-residue trans-membrane domain, and a 54-residue C-terminal cytoplasmic domain [wu et al]. The 19-residue TM domain forms the highly selective proton channel [Takashi et al]. Circular dichroism spectra has shown the TM domain to form one α-helix that spans the membrane [wu et al]. By analytical ultracentrifugation, the TM domain is found to form <scene name='User:Sarah_Henke/Sandbox_1/Alpha_hlix/1'>α-helical tetramers</scene>[takeuchi et al]. This tetrameric bundle of the TM domain is found by NMR to be tilted by 25-38° from the channel axis [takeuchi et al]. The trameric helices form a left-handed bundle that resembles a truncated cone <ref name="Stouffer" />. The TM helicies are arranged around the channel pore with an approximate fourfold rotational symmetry [takeuchi et al]. | The M2 proton channel from influenza A is 97 amino acid residues and forms a 24-residue N-terminal extracellular domain, a 19-residue trans-membrane domain, and a 54-residue C-terminal cytoplasmic domain [wu et al]. The 19-residue TM domain forms the highly selective proton channel [Takashi et al]. Circular dichroism spectra has shown the TM domain to form one α-helix that spans the membrane [wu et al]. By analytical ultracentrifugation, the TM domain is found to form <scene name='User:Sarah_Henke/Sandbox_1/Alpha_hlix/1'>α-helical tetramers</scene>[takeuchi et al]. This tetrameric bundle of the TM domain is found by NMR to be tilted by 25-38° from the channel axis [takeuchi et al]. The trameric helices form a left-handed bundle that resembles a truncated cone <ref name="Stouffer AL" />. The TM helicies are arranged around the channel pore with an approximate fourfold rotational symmetry [takeuchi et al]. | ||
== Central Cavity == | == Central Cavity == | ||
Revision as of 06:48, 30 September 2009
M2 Proton Channel from Influenza A VirusM2 Proton Channel from Influenza A Virus
|
BackgroundBackground
The M2 proton channel is a key protein that leads to viral infection [Takeuchi et al]. The M2 proton channel acidifies the viron which allows the viral matrix protein (M1) to disassociate from the ribonucleoprotein (RNP) [wu et al]. This allows the RNP to be transported to the nucleus of the cell [wu et al]. Several recent studies have looked at the effects of [1] and [Schnell et al] on inhibiting the transfer of protons through the M2 channel [2]. It has been found that M2 is resistant to these two drugs in 90% of humans, birds and pigs [2]. Understanding the structure and function of this proton channel is necessary in solving the resistance problem [3].
StructureStructure
The M2 proton channel from influenza A is 97 amino acid residues and forms a 24-residue N-terminal extracellular domain, a 19-residue trans-membrane domain, and a 54-residue C-terminal cytoplasmic domain [wu et al]. The 19-residue TM domain forms the highly selective proton channel [Takashi et al]. Circular dichroism spectra has shown the TM domain to form one α-helix that spans the membrane [wu et al]. By analytical ultracentrifugation, the TM domain is found to form [takeuchi et al]. This tetrameric bundle of the TM domain is found by NMR to be tilted by 25-38° from the channel axis [takeuchi et al]. The trameric helices form a left-handed bundle that resembles a truncated cone [3]. The TM helicies are arranged around the channel pore with an approximate fourfold rotational symmetry [takeuchi et al].
Central CavityCentral Cavity
|
pH GatingpH Gating
ReferencesReferences
- ↑ Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, Soto CS, Tereshko V, Nanda V, Stayrook S, DeGrado WF. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008 Jan 31;451(7178):596-9. PMID:18235504 doi:10.1038/nature06528
- ↑ 2.0 2.1 Cite error: Invalid
<ref>tag; no text was provided for refs namedStouffer - ↑ 3.0 3.1 Cite error: Invalid
<ref>tag; no text was provided for refs namedStouffer AL