1c05: Difference between revisions
New page: left|200px<br /><applet load="1c05" size="450" color="white" frame="true" align="right" spinBox="true" caption="1c05" /> '''SOLUTION STRUCTURE OF RIBOSOMAL PROTEIN S4 D... |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Image:1c05.gif|left|200px]]<br /><applet load="1c05" size=" | [[Image:1c05.gif|left|200px]]<br /><applet load="1c05" size="350" color="white" frame="true" align="right" spinBox="true" | ||
caption="1c05" /> | caption="1c05" /> | ||
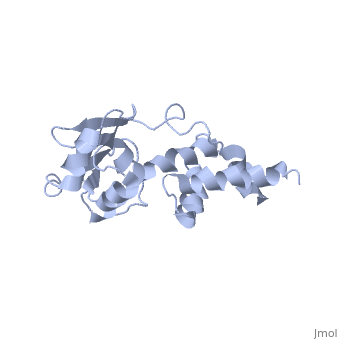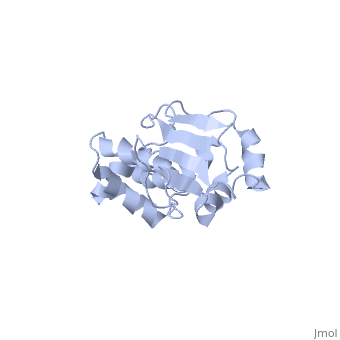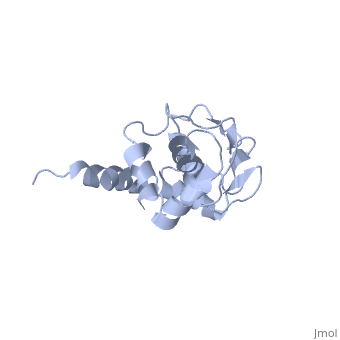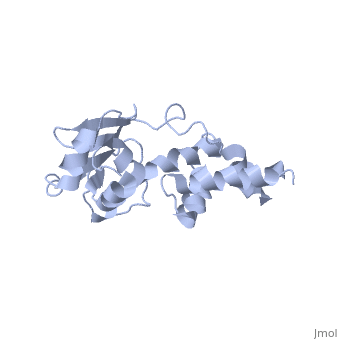
'''SOLUTION STRUCTURE OF RIBOSOMAL PROTEIN S4 DELTA 41, REFINED WITH DIPOLAR COUPLINGS (MINIMIZED AVERAGE STRUCTURE)'''<br /> | '''SOLUTION STRUCTURE OF RIBOSOMAL PROTEIN S4 DELTA 41, REFINED WITH DIPOLAR COUPLINGS (MINIMIZED AVERAGE STRUCTURE)'''<br /> | ||
==Overview== | ==Overview== | ||
Prokaryotic protein S4 initiates assembly of the small ribosomal subunit | Prokaryotic protein S4 initiates assembly of the small ribosomal subunit by binding to 16 S rRNA. Residues 43-200 of S4 from Bacillus stearothermophilus (S4 Delta41) bind to both 16 S rRNA and to a mRNA pseudoknot. In order to obtain structure-based insights regarding RNA binding, we previously determined the solution structure of S4 Delta41 using NOE, hydrogen bond, and torsion angle restraints. S4 Delta41 is elongated, with two distinct subdomains, one all helical, the other including a beta-sheet. In contrast to the high resolution structures obtained for each individual subdomain, their relative orientation was not precisely defined because only 17 intersubdomain NOE restraints were determined. Compared to the 1.7 A crystal structure, when the sheet-containing subdomains are superimposed, the helical subdomain is twisted by almost 45 degrees about the long axis of the molecule in the solution structure. Because variations in subdomain orientation may explain how the protein recognizes multiple RNA targets, our current goal is to determine the orientation of the subdomains in solution with high precision. To this end, NOE assignments were re-examined. NOESY experiments on a specifically labeled sample revealed that one of the intersubdomain restraints had been misassigned. However, the revised set of NOE restraints produces solution structures that still have imprecisely defined subdomain orientations and that lie between the original NMR structure and the crystal structure. In contrast, augmenting the NOE restraints with N-H dipolar couplings, measured in uniaxial liquid crystalline phases, clearly establishes the relative orientation of the subdomains. Data obtained from two independent liquid crystalline milieux, DMPC/DHPC bicelles and the filamentous bacteriophage Pf1, show that the relative orientation of the subdomains in solution is quite similar to the subdomain orientation in the crystal structure. The solution structure, refined with dipolar data, is presented and its implications for S4's RNA binding activity are discussed. | ||
==About this Structure== | ==About this Structure== | ||
1C05 is a [http://en.wikipedia.org/wiki/Single_protein Single protein] structure of sequence from [http://en.wikipedia.org/wiki/Geobacillus_stearothermophilus Geobacillus stearothermophilus]. Full crystallographic information is available from [http:// | 1C05 is a [http://en.wikipedia.org/wiki/Single_protein Single protein] structure of sequence from [http://en.wikipedia.org/wiki/Geobacillus_stearothermophilus Geobacillus stearothermophilus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1C05 OCA]. | ||
==Reference== | ==Reference== | ||
| Line 13: | Line 13: | ||
[[Category: Geobacillus stearothermophilus]] | [[Category: Geobacillus stearothermophilus]] | ||
[[Category: Single protein]] | [[Category: Single protein]] | ||
[[Category: Draper, D | [[Category: Draper, D E.]] | ||
[[Category: Gerstner, R | [[Category: Gerstner, R B.]] | ||
[[Category: Markus, M | [[Category: Markus, M A.]] | ||
[[Category: Torchia, D | [[Category: Torchia, D A.]] | ||
[[Category: possible helix-turn-helix motif]] | [[Category: possible helix-turn-helix motif]] | ||
[[Category: two subdomains]] | [[Category: two subdomains]] | ||
[[Category: unique topology]] | [[Category: unique topology]] | ||
''Page seeded by [http:// | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Feb 21 12:01:01 2008'' | ||
Revision as of 13:01, 21 February 2008
|
SOLUTION STRUCTURE OF RIBOSOMAL PROTEIN S4 DELTA 41, REFINED WITH DIPOLAR COUPLINGS (MINIMIZED AVERAGE STRUCTURE)
OverviewOverview
Prokaryotic protein S4 initiates assembly of the small ribosomal subunit by binding to 16 S rRNA. Residues 43-200 of S4 from Bacillus stearothermophilus (S4 Delta41) bind to both 16 S rRNA and to a mRNA pseudoknot. In order to obtain structure-based insights regarding RNA binding, we previously determined the solution structure of S4 Delta41 using NOE, hydrogen bond, and torsion angle restraints. S4 Delta41 is elongated, with two distinct subdomains, one all helical, the other including a beta-sheet. In contrast to the high resolution structures obtained for each individual subdomain, their relative orientation was not precisely defined because only 17 intersubdomain NOE restraints were determined. Compared to the 1.7 A crystal structure, when the sheet-containing subdomains are superimposed, the helical subdomain is twisted by almost 45 degrees about the long axis of the molecule in the solution structure. Because variations in subdomain orientation may explain how the protein recognizes multiple RNA targets, our current goal is to determine the orientation of the subdomains in solution with high precision. To this end, NOE assignments were re-examined. NOESY experiments on a specifically labeled sample revealed that one of the intersubdomain restraints had been misassigned. However, the revised set of NOE restraints produces solution structures that still have imprecisely defined subdomain orientations and that lie between the original NMR structure and the crystal structure. In contrast, augmenting the NOE restraints with N-H dipolar couplings, measured in uniaxial liquid crystalline phases, clearly establishes the relative orientation of the subdomains. Data obtained from two independent liquid crystalline milieux, DMPC/DHPC bicelles and the filamentous bacteriophage Pf1, show that the relative orientation of the subdomains in solution is quite similar to the subdomain orientation in the crystal structure. The solution structure, refined with dipolar data, is presented and its implications for S4's RNA binding activity are discussed.
About this StructureAbout this Structure
1C05 is a Single protein structure of sequence from Geobacillus stearothermophilus. Full crystallographic information is available from OCA.
ReferenceReference
Refining the overall structure and subdomain orientation of ribosomal protein S4 delta41 with dipolar couplings measured by NMR in uniaxial liquid crystalline phases., Markus MA, Gerstner RB, Draper DE, Torchia DA, J Mol Biol. 1999 Sep 17;292(2):375-87. PMID:10493882
Page seeded by OCA on Thu Feb 21 12:01:01 2008