3pmq: Difference between revisions
No edit summary |
No edit summary |
||
| Line 3: | Line 3: | ||
<StructureSection load='3pmq' size='340' side='right'caption='[[3pmq]], [[Resolution|resolution]] 3.20Å' scene=''> | <StructureSection load='3pmq' size='340' side='right'caption='[[3pmq]], [[Resolution|resolution]] 3.20Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[3pmq]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/ | <table><tr><td colspan='2'>[[3pmq]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Shewanella_oneidensis_MR-1 Shewanella oneidensis MR-1]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3PMQ OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3PMQ FirstGlance]. <br> | ||
</td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=CA:CALCIUM+ION'>CA</scene>, <scene name='pdbligand=HEC:HEME+C'>HEC</scene></td></tr> | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=CA:CALCIUM+ION'>CA</scene>, <scene name='pdbligand=HEC:HEME+C'>HEC</scene></td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3pmq FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3pmq OCA], [https://pdbe.org/3pmq PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3pmq RCSB], [https://www.ebi.ac.uk/pdbsum/3pmq PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3pmq ProSAT]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3pmq FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3pmq OCA], [https://pdbe.org/3pmq PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3pmq RCSB], [https://www.ebi.ac.uk/pdbsum/3pmq PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3pmq ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | |||
[https://www.uniprot.org/uniprot/Q8EG32_SHEON Q8EG32_SHEON] | |||
<div style="background-color:#fffaf0;"> | <div style="background-color:#fffaf0;"> | ||
== Publication Abstract from PubMed == | == Publication Abstract from PubMed == | ||
| Line 24: | Line 26: | ||
</StructureSection> | </StructureSection> | ||
[[Category: Large Structures]] | [[Category: Large Structures]] | ||
[[Category: Shewanella oneidensis]] | [[Category: Shewanella oneidensis MR-1]] | ||
[[Category: Clarke | [[Category: Clarke TA]] | ||
[[Category: Edwards | [[Category: Edwards MJ]] | ||
[[Category: Richardson | [[Category: Richardson DJ]] | ||
Revision as of 08:48, 31 May 2023
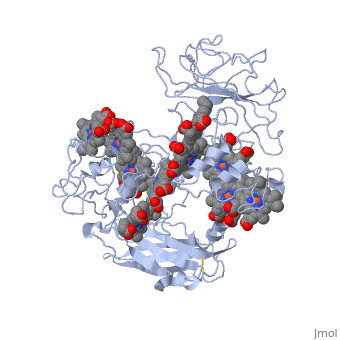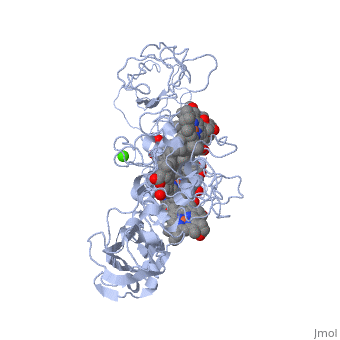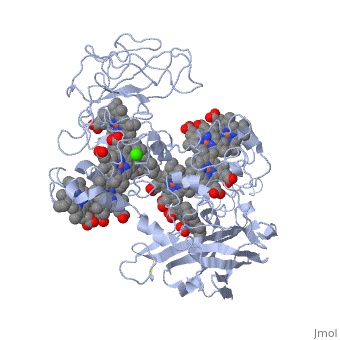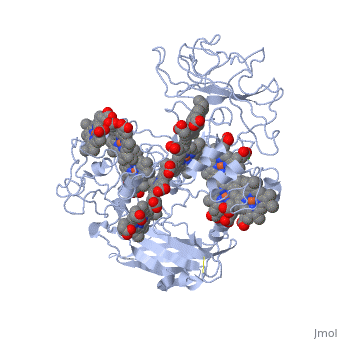
Crystal structure of the outer membrane decaheme cytochrome MtrFCrystal structure of the outer membrane decaheme cytochrome MtrF
Structural highlights
FunctionPublication Abstract from PubMedSome bacterial species are able to utilize extracellular mineral forms of iron and manganese as respiratory electron acceptors. In Shewanella oneidensis this involves decaheme cytochromes that are located on the bacterial cell surface at the termini of trans-outer-membrane electron transfer conduits. The cell surface cytochromes can potentially play multiple roles in mediating electron transfer directly to insoluble electron sinks, catalyzing electron exchange with flavin electron shuttles or participating in extracellular intercytochrome electron exchange along "nanowire" appendages. We present a 3.2-A crystal structure of one of these decaheme cytochromes, MtrF, that allows the spatial organization of the 10 hemes to be visualized for the first time. The hemes are organized across four domains in a unique crossed conformation, in which a staggered 65-A octaheme chain transects the length of the protein and is bisected by a planar 45-A tetraheme chain that connects two extended Greek key split beta-barrel domains. The structure provides molecular insight into how reduction of insoluble substrate (e.g., minerals), soluble substrates (e.g., flavins), and cytochrome redox partners might be possible in tandem at different termini of a trifurcated electron transport chain on the cell surface. Structure of a bacterial cell surface decaheme electron conduit.,Clarke TA, Edwards MJ, Gates AJ, Hall A, White GF, Bradley J, Reardon CL, Shi L, Beliaev AS, Marshall MJ, Wang Z, Watmough NJ, Fredrickson JK, Zachara JM, Butt JN, Richardson DJ Proc Natl Acad Sci U S A. 2011 May 23. PMID:21606337[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||