Major histocompatibility complex: Difference between revisions
No edit summary |
No edit summary |
||
| Line 4: | Line 4: | ||
''' Major Histocompatibility Complex''' (MHC) molecules bind peptides derived from degraded proteins and present these peptides on the surface of the cell. Cytotoxic T-lymphocytes or helper T cells recognize the MHC:peptide complex on the surface of the cell and trigger an immune response if the presented peptide (antigen) is suggestive of a pathogenic or foreign protein. In this way, MHC molecules allow for immune system detection of protein presence, making them an essential part of the organism’s immune system. MHC molecules are divided into Class I and Class II molecules based on the types of cells that typically express them and the types of peptides they typically bind. | ''' Major Histocompatibility Complex''' (MHC) molecules bind peptides derived from degraded proteins and present these peptides on the surface of the cell. Cytotoxic T-lymphocytes or helper T cells recognize the MHC:peptide complex on the surface of the cell and trigger an immune response if the presented peptide (antigen) is suggestive of a pathogenic or foreign protein. In this way, MHC molecules allow for immune system detection of protein presence, making them an essential part of the organism’s immune system. MHC molecules are divided into Class I and Class II molecules based on the types of cells that typically express them and the types of peptides they typically bind. | ||
<br /> | <br /> | ||
* '''MHC class I''' ([[Major Histocompatibility Complex Class I]]) are found in all nucleated cells. These cell surface proteins display peptides from cellular intrinsic proteins<ref>PMID:18675588</ref>. For more details see <br /> | * '''MHC class I''' ([[Major Histocompatibility Complex Class I]]) are found in all nucleated cells and platelets. These cell surface proteins display peptides from cellular intrinsic proteins<ref>PMID:18675588</ref>. For more details see <br /> | ||
**[[Effect of HCMV on Major Histocompatibility Complex Class I]]<br /> | **[[Effect of HCMV on Major Histocompatibility Complex Class I]]<br /> | ||
**[[MR1 Binds Vitamin Metabolites]].<br /> | **[[MR1 Binds Vitamin Metabolites]].<br /> | ||
| Line 11: | Line 11: | ||
== Structural highlights == | == Structural highlights == | ||
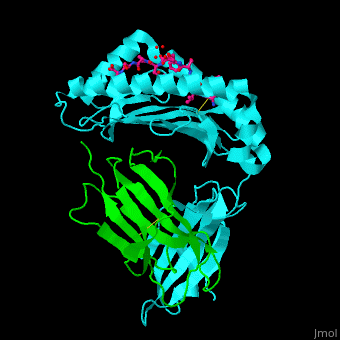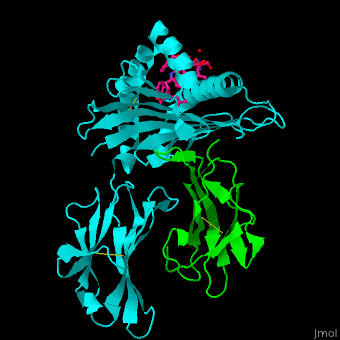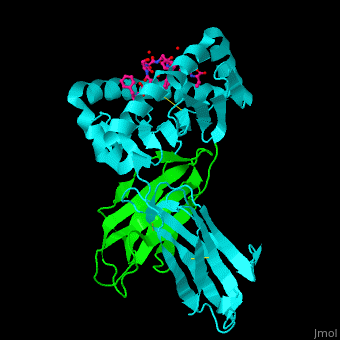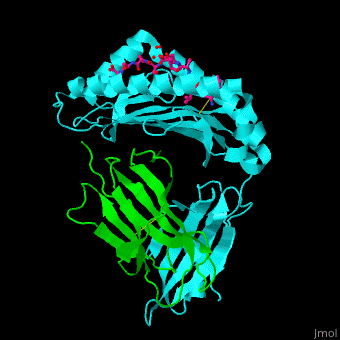
Both Class I and Class II MHC molecules are heterodimers with two extracellular subunits ( and and one or two transmembrane helices that extend from the extracellular subunits to the cytoplasm. In Class I molecules, the subunit is divided into three domains (and). The anddomains form a -sheet platform with two -helix rails that serve as the peptide-binding groove. The domain forms an immunoglobin-like fold that carries the peptide-binding groove with added support from with the subunit (a 2-microglobulin molecule encoded outside of the MHC Class I gene locus). In Class II molecules, both the and subunits are divided into two domains (and). The peptide-binding groove is formed by the and domains and the and domains carry the peptide-binding groove. While the subunit is polymorphic for both MHC Classes, the subunit is polymorphic only for Class II molecules. | |||
<scene name='45/457390/Cv/5'>Human MHC class I antigen with β 2-microglobulin and peptide from Hepatitis virus</scene>. | <scene name='45/457390/Cv/5'>Human MHC class I antigen with β 2-microglobulin and peptide from Hepatitis virus</scene>. | ||
The <scene name='45/457390/Cv/6'>peptide derived from Hepatitis virus binds MHC in a peptide-recognition groove and makes various interactions with side chains and with water molecules</scene><ref>PMID:21538979</ref>. Water molecules shown as red spheres. | The <scene name='45/457390/Cv/6'>peptide derived from Hepatitis virus binds MHC in a peptide-recognition groove and makes various interactions with side chains and with water molecules</scene><ref>PMID:21538979</ref>. Water molecules shown as red spheres. | ||
==3D | ==List of Published 3D Structures of MHC== | ||
[[MHC 3D structures of MHC]] | [[MHC 3D structures of MHC]] | ||
</StructureSection> | </StructureSection> | ||
Revision as of 00:03, 22 July 2020
FunctionMajor Histocompatibility Complex (MHC) molecules bind peptides derived from degraded proteins and present these peptides on the surface of the cell. Cytotoxic T-lymphocytes or helper T cells recognize the MHC:peptide complex on the surface of the cell and trigger an immune response if the presented peptide (antigen) is suggestive of a pathogenic or foreign protein. In this way, MHC molecules allow for immune system detection of protein presence, making them an essential part of the organism’s immune system. MHC molecules are divided into Class I and Class II molecules based on the types of cells that typically express them and the types of peptides they typically bind.
For information on MHC class II interactions with T-cell receptor and gliadin peptide see SP3.4-TCR-HLA-DQ8-α-1-gliadin complex. Structural highlightsBoth Class I and Class II MHC molecules are heterodimers with two extracellular subunits ( and and one or two transmembrane helices that extend from the extracellular subunits to the cytoplasm. In Class I molecules, the subunit is divided into three domains (and). The anddomains form a -sheet platform with two -helix rails that serve as the peptide-binding groove. The domain forms an immunoglobin-like fold that carries the peptide-binding groove with added support from with the subunit (a 2-microglobulin molecule encoded outside of the MHC Class I gene locus). In Class II molecules, both the and subunits are divided into two domains (and). The peptide-binding groove is formed by the and domains and the and domains carry the peptide-binding groove. While the subunit is polymorphic for both MHC Classes, the subunit is polymorphic only for Class II molecules. . The [3]. Water molecules shown as red spheres. List of Published 3D Structures of MHC |
| ||||||||||
ReferencesReferences
- ↑ Raghavan M, Del Cid N, Rizvi SM, Peters LR. MHC class I assembly: out and about. Trends Immunol. 2008 Sep;29(9):436-43. doi: 10.1016/j.it.2008.06.004. PMID:18675588 doi:http://dx.doi.org/10.1016/j.it.2008.06.004
- ↑ Holling TM, Schooten E, van Den Elsen PJ. Function and regulation of MHC class II molecules in T-lymphocytes: of mice and men. Hum Immunol. 2004 Apr;65(4):282-90. PMID:15120183 doi:http://dx.doi.org/10.1016/j.humimm.2004.01.005
- ↑ Liu J, Chen KY, Ren EC. Structural insights into the binding of hepatitis B virus core peptide to HLA-A2 alleles: Towards designing better vaccines. Eur J Immunol. 2011 Jul;41(7):2097-106. doi: 10.1002/eji.201041370. PMID:21538979 doi:10.1002/eji.201041370