1e9z: Difference between revisions
No edit summary |
No edit summary |
||
| Line 3: | Line 3: | ||
<StructureSection load='1e9z' size='340' side='right'caption='[[1e9z]], [[Resolution|resolution]] 3.00Å' scene=''> | <StructureSection load='1e9z' size='340' side='right'caption='[[1e9z]], [[Resolution|resolution]] 3.00Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[1e9z]] is a 2 chain structure with sequence from [ | <table><tr><td colspan='2'>[[1e9z]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Atcc_43504 Atcc 43504]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1E9Z OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1E9Z FirstGlance]. <br> | ||
</td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=NI:NICKEL+(II)+ION'>NI</scene></td></tr> | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=NI:NICKEL+(II)+ION'>NI</scene></td></tr> | ||
<tr id='NonStdRes'><td class="sblockLbl"><b>[[Non-Standard_Residue|NonStd Res:]]</b></td><td class="sblockDat"><scene name='pdbligand=KCX:LYSINE+NZ-CARBOXYLIC+ACID'>KCX</scene></td></tr> | <tr id='NonStdRes'><td class="sblockLbl"><b>[[Non-Standard_Residue|NonStd Res:]]</b></td><td class="sblockDat"><scene name='pdbligand=KCX:LYSINE+NZ-CARBOXYLIC+ACID'>KCX</scene></td></tr> | ||
<tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[1e9y|1e9y]]</td></tr> | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat"><div style='overflow: auto; max-height: 3em;'>[[1e9y|1e9y]]</div></td></tr> | ||
<tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[ | <tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[https://en.wikipedia.org/wiki/Urease Urease], with EC number [https://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.5.1.5 3.5.1.5] </span></td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1e9z FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1e9z OCA], [https://pdbe.org/1e9z PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1e9z RCSB], [https://www.ebi.ac.uk/pdbsum/1e9z PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1e9z ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
[[ | [[https://www.uniprot.org/uniprot/URE23_HELPY URE23_HELPY]] Ammonia produced by ureolysis increases the gastric pH thereby providing an environment permissive for colonization of the stomach.<ref>PMID:8039935</ref> [[https://www.uniprot.org/uniprot/URE1_HELPY URE1_HELPY]] Ammonia produced by ureolysis increases the gastric pH thereby providing an environment permissive for colonization of the stomach.<ref>PMID:8039935</ref> | ||
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 34: | Line 34: | ||
==See Also== | ==See Also== | ||
*[[Urease|Urease]] | *[[Urease|Urease]] | ||
*[[Urease | *[[Urease 3D structures|Urease 3D structures]] | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 16:11, 13 October 2021
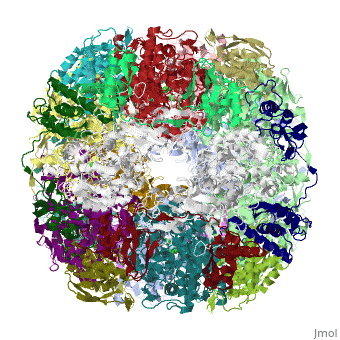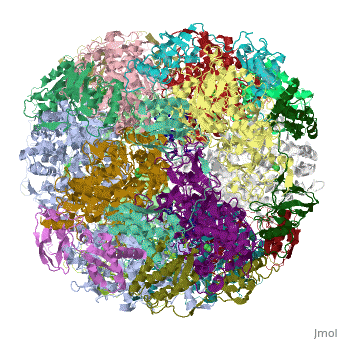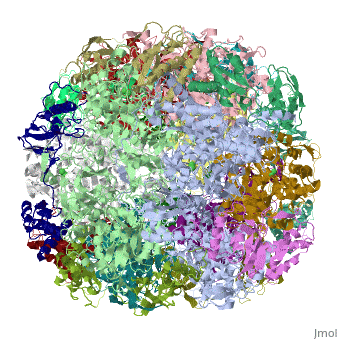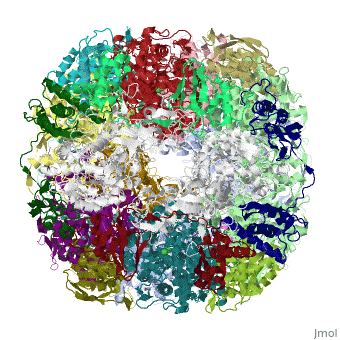
Crystal structure of Helicobacter pylori ureaseCrystal structure of Helicobacter pylori urease
Structural highlights
Function[URE23_HELPY] Ammonia produced by ureolysis increases the gastric pH thereby providing an environment permissive for colonization of the stomach.[1] [URE1_HELPY] Ammonia produced by ureolysis increases the gastric pH thereby providing an environment permissive for colonization of the stomach.[2] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedHelicobacter pylori, an etiologic agent in a variety of gastroduodenal diseases, produces a large amount of urease, which is believed to neutralize gastric acid by producing ammonia for the survival of the bacteria. Up to 30% of the enzyme associates with the surface of intact cells upon lysis of neighboring bacteria. The role of the enzyme at the extracellular location has been a subject of controversy because the purified enzyme is irreversibly inactivated below pH 5. We have determined the crystal structure of H. pylori urease, which has a 1.1 MDa spherical assembly of 12 catalytic units with an outer diameter of approximately 160 A. Under physiologically relevant conditions, the activity of the enzyme remains unaffected down to pH 3. Activity assays under different conditions indicated that the cluster of the 12 active sites on the supramolecular assembly may be critical for the survival of the enzyme at low pH. The structure provides a novel example of a molecular assembly adapted for acid resistance that, together with the low Km value of the enzyme, is likely to enable the organism to inhabit the hostile niche. Supramolecular assembly and acid resistance of Helicobacter pylori urease.,Ha NC, Oh ST, Sung JY, Cha KA, Lee MH, Oh BH Nat Struct Biol. 2001 Jun;8(6):505-9. PMID:11373617[3] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||||||