3es9: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
==NADPH-Cytochrome P450 Reductase in an Open Conformation== | ==NADPH-Cytochrome P450 Reductase in an Open Conformation== | ||
<StructureSection load='3es9' size='340' side='right' caption='[[3es9]], [[Resolution|resolution]] 3.40Å' scene=''> | <StructureSection load='3es9' size='340' side='right'caption='[[3es9]], [[Resolution|resolution]] 3.40Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[3es9]] is a 3 chain structure with sequence from [ | <table><tr><td colspan='2'>[[3es9]] is a 3 chain structure with sequence from [https://en.wikipedia.org/wiki/Buffalo_rat Buffalo rat]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3ES9 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3ES9 FirstGlance]. <br> | ||
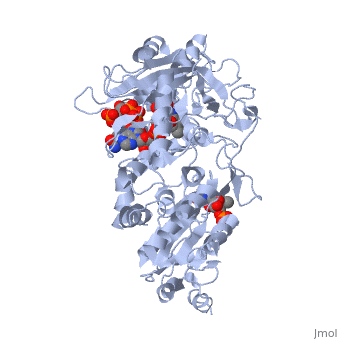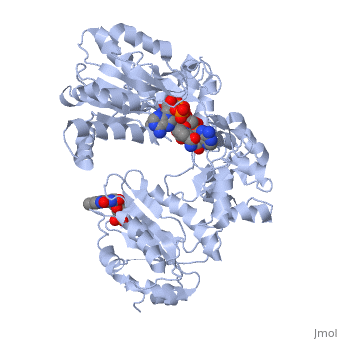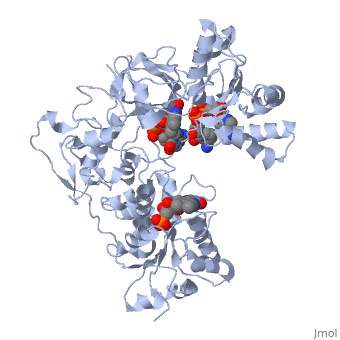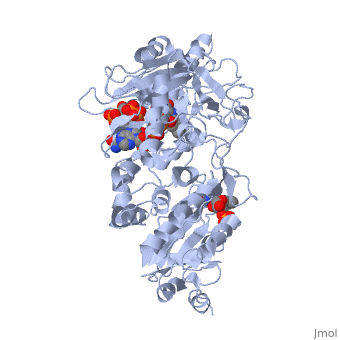
</td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=FAD:FLAVIN-ADENINE+DINUCLEOTIDE'>FAD</scene>, <scene name='pdbligand=FMN:FLAVIN+MONONUCLEOTIDE'>FMN</scene>, <scene name='pdbligand=NAP:NADP+NICOTINAMIDE-ADENINE-DINUCLEOTIDE+PHOSPHATE'>NAP</scene></td></tr> | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=FAD:FLAVIN-ADENINE+DINUCLEOTIDE'>FAD</scene>, <scene name='pdbligand=FMN:FLAVIN+MONONUCLEOTIDE'>FMN</scene>, <scene name='pdbligand=NAP:NADP+NICOTINAMIDE-ADENINE-DINUCLEOTIDE+PHOSPHATE'>NAP</scene></td></tr> | ||
<tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">CYPOR, Por ([ | <tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">CYPOR, Por ([https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=10116 Buffalo rat])</td></tr> | ||
<tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[ | <tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[https://en.wikipedia.org/wiki/NADPH--hemoprotein_reductase NADPH--hemoprotein reductase], with EC number [https://www.brenda-enzymes.info/php/result_flat.php4?ecno=1.6.2.4 1.6.2.4] </span></td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3es9 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3es9 OCA], [https://pdbe.org/3es9 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3es9 RCSB], [https://www.ebi.ac.uk/pdbsum/3es9 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3es9 ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
[[ | [[https://www.uniprot.org/uniprot/NCPR_RAT NCPR_RAT]] This enzyme is required for electron transfer from NADP to cytochrome P450 in microsomes. It can also provide electron transfer to heme oxygenase and cytochrome B5. | ||
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 32: | Line 32: | ||
==See Also== | ==See Also== | ||
*[[NADPH-Cytochrome P450 Reductase|NADPH-Cytochrome P450 Reductase]] | *[[NADPH-Cytochrome P450 Reductase|NADPH-Cytochrome P450 Reductase]] | ||
== References == | == References == | ||
| Line 39: | Line 38: | ||
</StructureSection> | </StructureSection> | ||
[[Category: Buffalo rat]] | [[Category: Buffalo rat]] | ||
[[Category: Large Structures]] | |||
[[Category: NADPH--hemoprotein reductase]] | [[Category: NADPH--hemoprotein reductase]] | ||
[[Category: Hamdane, D]] | [[Category: Hamdane, D]] | ||
Revision as of 12:14, 23 February 2022
NADPH-Cytochrome P450 Reductase in an Open ConformationNADPH-Cytochrome P450 Reductase in an Open Conformation
Structural highlights
Function[NCPR_RAT] This enzyme is required for electron transfer from NADP to cytochrome P450 in microsomes. It can also provide electron transfer to heme oxygenase and cytochrome B5. Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedNADPH-cytochrome P450 oxidoreductase (CYPOR) catalyzes the transfer of electrons to all known microsomal cytochromes P450. A CYPOR variant, with a 4-amino acid deletion in the hinge connecting the FMN domain to the rest of the protein, has been crystallized in three remarkably extended conformations. The variant donates an electron to cytochrome P450 at the same rate as the wild-type, when provided with sufficient electrons. Nevertheless, it is defective in its ability to transfer electrons intramolecularly from FAD to FMN. The three extended CYPOR structures demonstrate that, by pivoting on the C terminus of the hinge, the FMN domain of the enzyme undergoes a structural rearrangement that separates it from FAD and exposes the FMN, allowing it to interact with its redox partners. A similar movement most likely occurs in the wild-type enzyme in the course of transferring electrons from FAD to its physiological partner, cytochrome P450. A model of the complex between an open conformation of CYPOR and cytochrome P450 is presented that satisfies mutagenesis constraints. Neither lengthening the linker nor mutating its sequence influenced the activity of CYPOR. It is likely that the analogous linker in other members of the diflavin family functions in a similar manner. Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450.,Hamdane D, Xia C, Im SC, Zhang H, Kim JJ, Waskell L J Biol Chem. 2009 Apr 24;284(17):11374-84. Epub 2009 Jan 26. PMID:19171935[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||||