1mq8: Difference between revisions
No edit summary |
No edit summary |
||
| Line 4: | Line 4: | ||
|PDB= 1mq8 |SIZE=350|CAPTION= <scene name='initialview01'>1mq8</scene>, resolution 3.30Å | |PDB= 1mq8 |SIZE=350|CAPTION= <scene name='initialview01'>1mq8</scene>, resolution 3.30Å | ||
|SITE= | |SITE= | ||
|LIGAND= <scene name='pdbligand= | |LIGAND= <scene name='pdbligand=MG:MAGNESIUM+ION'>MG</scene>, <scene name='pdbligand=NAG:N-ACETYL-D-GLUCOSAMINE'>NAG</scene> | ||
|ACTIVITY= | |ACTIVITY= | ||
|GENE= ICAM-1 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=9606 Homo sapiens]), LFA-1 (AlphaLbeta2) ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=9606 Homo sapiens]) | |GENE= ICAM-1 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=9606 Homo sapiens]), LFA-1 (AlphaLbeta2) ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=9606 Homo sapiens]) | ||
|DOMAIN= | |||
|RELATEDENTRY=[[1mq9|1MQ9]], [[1mqa|1MQA]] | |||
|RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1mq8 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1mq8 OCA], [http://www.ebi.ac.uk/pdbsum/1mq8 PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=1mq8 RCSB]</span> | |||
}} | }} | ||
| Line 14: | Line 17: | ||
==Overview== | ==Overview== | ||
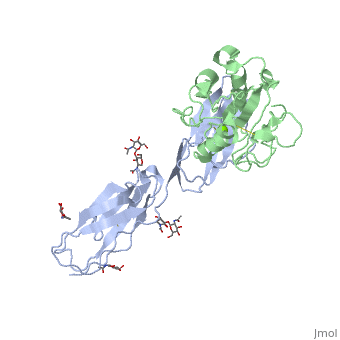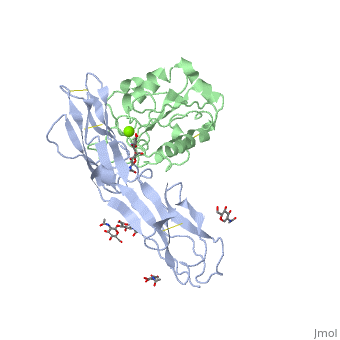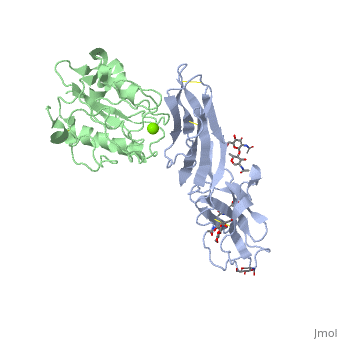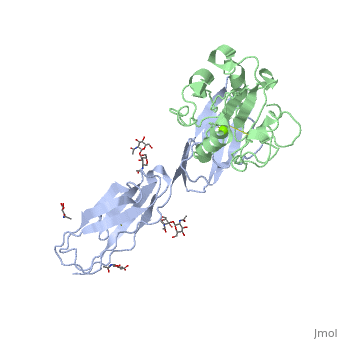
The structure of the I domain of integrin alpha L beta 2 bound to the Ig superfamily ligand ICAM-1 reveals the open ligand binding conformation and the first example of an integrin-IgSF interface. The I domain Mg2+ directly coordinates Glu-34 of ICAM-1, and a dramatic swing of I domain residue Glu-241 enables a critical salt bridge. Liganded and unliganded structures for both high- and intermediate-affinity mutant I domains reveal that ligand binding can induce conformational change in the alpha L I domain and that allosteric signals can convert the closed conformation to intermediate or open conformations without ligand binding. Pulling down on the C-terminal alpha 7 helix with introduced disulfide bonds ratchets the beta 6-alpha 7 loop into three different positions in the closed, intermediate, and open conformations, with a progressive increase in affinity. | The structure of the I domain of integrin alpha L beta 2 bound to the Ig superfamily ligand ICAM-1 reveals the open ligand binding conformation and the first example of an integrin-IgSF interface. The I domain Mg2+ directly coordinates Glu-34 of ICAM-1, and a dramatic swing of I domain residue Glu-241 enables a critical salt bridge. Liganded and unliganded structures for both high- and intermediate-affinity mutant I domains reveal that ligand binding can induce conformational change in the alpha L I domain and that allosteric signals can convert the closed conformation to intermediate or open conformations without ligand binding. Pulling down on the C-terminal alpha 7 helix with introduced disulfide bonds ratchets the beta 6-alpha 7 loop into three different positions in the closed, intermediate, and open conformations, with a progressive increase in affinity. | ||
==About this Structure== | ==About this Structure== | ||
| Line 37: | Line 37: | ||
[[Category: Yang, Y.]] | [[Category: Yang, Y.]] | ||
[[Category: Zhang, R.]] | [[Category: Zhang, R.]] | ||
[[Category: ig superfamily]] | [[Category: ig superfamily]] | ||
[[Category: metal mediated protein interface]] | [[Category: metal mediated protein interface]] | ||
[[Category: rossmann fold]] | [[Category: rossmann fold]] | ||
''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Sun Mar 30 22:19:38 2008'' | ||
Revision as of 22:19, 30 March 2008
| |||||||
| , resolution 3.30Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||
| Gene: | ICAM-1 (Homo sapiens), LFA-1 (AlphaLbeta2) (Homo sapiens) | ||||||
| Related: | 1MQ9, 1MQA
| ||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
Crystal structure of alphaL I domain in complex with ICAM-1
OverviewOverview
The structure of the I domain of integrin alpha L beta 2 bound to the Ig superfamily ligand ICAM-1 reveals the open ligand binding conformation and the first example of an integrin-IgSF interface. The I domain Mg2+ directly coordinates Glu-34 of ICAM-1, and a dramatic swing of I domain residue Glu-241 enables a critical salt bridge. Liganded and unliganded structures for both high- and intermediate-affinity mutant I domains reveal that ligand binding can induce conformational change in the alpha L I domain and that allosteric signals can convert the closed conformation to intermediate or open conformations without ligand binding. Pulling down on the C-terminal alpha 7 helix with introduced disulfide bonds ratchets the beta 6-alpha 7 loop into three different positions in the closed, intermediate, and open conformations, with a progressive increase in affinity.
About this StructureAbout this Structure
1MQ8 is a Protein complex structure of sequences from Homo sapiens. Full crystallographic information is available from OCA.
ReferenceReference
Structures of the alpha L I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation., Shimaoka M, Xiao T, Liu JH, Yang Y, Dong Y, Jun CD, McCormack A, Zhang R, Joachimiak A, Takagi J, Wang JH, Springer TA, Cell. 2003 Jan 10;112(1):99-111. PMID:12526797
Page seeded by OCA on Sun Mar 30 22:19:38 2008