1jfp: Difference between revisions
No edit summary |
No edit summary |
||
| Line 3: | Line 3: | ||
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[1jfp]] is a 1 chain structure with sequence from [http://en.wikipedia.org/wiki/Bos_taurus Bos taurus]. Full experimental information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1JFP OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1JFP FirstGlance]. <br> | <table><tr><td colspan='2'>[[1jfp]] is a 1 chain structure with sequence from [http://en.wikipedia.org/wiki/Bos_taurus Bos taurus]. Full experimental information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1JFP OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1JFP FirstGlance]. <br> | ||
</td></tr><tr><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=RET:RETINAL'>RET</scene>< | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=RET:RETINAL'>RET</scene></td></tr> | ||
<tr><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[1eds|1eds]], [[1edv|1edv]], [[1edw|1edw]], [[1edx|1edx]], [[1fdf|1fdf]]</td></tr> | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[1eds|1eds]], [[1edv|1edv]], [[1edw|1edw]], [[1edx|1edx]], [[1fdf|1fdf]]</td></tr> | ||
<tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1jfp FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1jfp OCA], [http://www.rcsb.org/pdb/explore.do?structureId=1jfp RCSB], [http://www.ebi.ac.uk/pdbsum/1jfp PDBsum]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1jfp FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1jfp OCA], [http://www.rcsb.org/pdb/explore.do?structureId=1jfp RCSB], [http://www.ebi.ac.uk/pdbsum/1jfp PDBsum]</span></td></tr> | ||
<table> | </table> | ||
== Function == | |||
[[http://www.uniprot.org/uniprot/OPSD_BOVIN OPSD_BOVIN]] Photoreceptor required for image-forming vision at low light intensity. Required for photoreceptor cell viability after birth. Light-induced isomerization of 11-cis to all-trans retinal triggers a conformational change leading to G-protein activation and release of all-trans retinal (By similarity).<ref>PMID:16908857</ref> <ref>PMID:17060607</ref> | |||
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 33: | Line 35: | ||
</StructureSection> | </StructureSection> | ||
[[Category: Bos taurus]] | [[Category: Bos taurus]] | ||
[[Category: Albert, A D | [[Category: Albert, A D]] | ||
[[Category: Choi, G | [[Category: Choi, G]] | ||
[[Category: Yeagle, P L | [[Category: Yeagle, P L]] | ||
[[Category: G-protein coupled receptor]] | [[Category: G-protein coupled receptor]] | ||
[[Category: Membrane protein]] | [[Category: Membrane protein]] | ||
[[Category: Signaling protein]] | [[Category: Signaling protein]] | ||
Revision as of 12:05, 25 December 2014
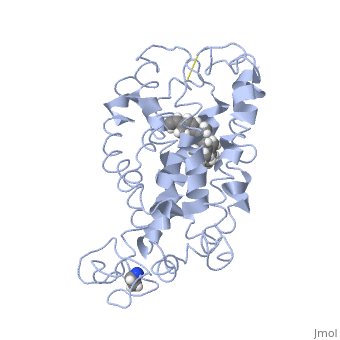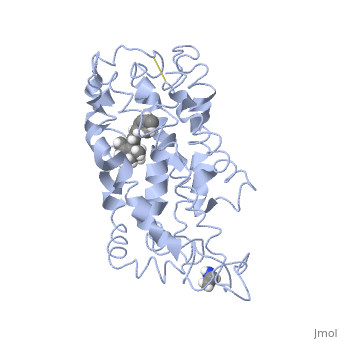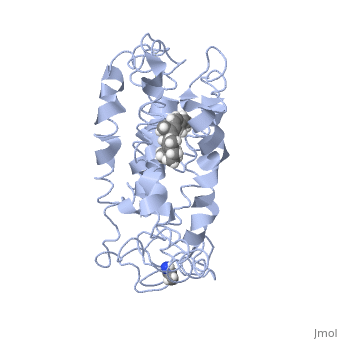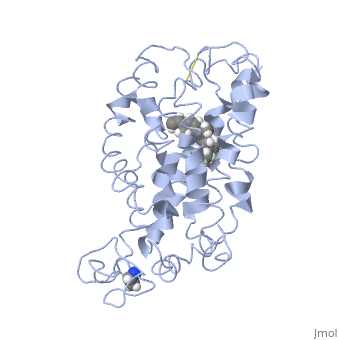
Structure of bovine rhodopsin (dark adapted)Structure of bovine rhodopsin (dark adapted)
Structural highlights
Function[OPSD_BOVIN] Photoreceptor required for image-forming vision at low light intensity. Required for photoreceptor cell viability after birth. Light-induced isomerization of 11-cis to all-trans retinal triggers a conformational change leading to G-protein activation and release of all-trans retinal (By similarity).[1] [2] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedActivation of G-protein coupled receptors (GPCR) is not yet understood. A recent structure showed most of rhodopsin in the ground (not activated) state of the GPCR, but the cytoplasmic face, which couples to the G protein in signal transduction, was not well-defined. We have determined an experimental three-dimensional structure for rhodopsin in the unactivated state, which shows good agreement with the crystal structure in the transmembrane domain. This new structure defines the cytoplasmic face of rhodopsin. The G-protein binding site can be mapped. The same experimental approach yields a preliminary structure of the cytoplasmic face in the activated (metarhodopsin II) receptor. Differences between the two structures suggest how the receptor is activated to couple with transducin. Studies on the structure of the G-protein-coupled receptor rhodopsin including the putative G-protein binding site in unactivated and activated forms.,Yeagle PL, Choi G, Albert AD Biochemistry. 2001 Oct 2;40(39):11932-7. PMID:11570894[3] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||