1a3n: Difference between revisions
No edit summary |
No edit summary |
||
| Line 4: | Line 4: | ||
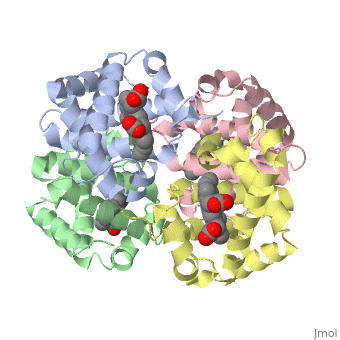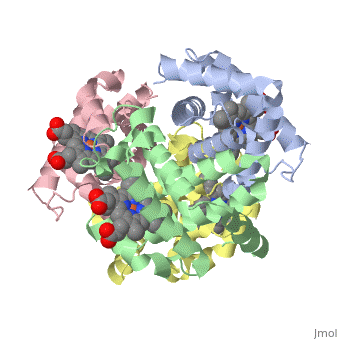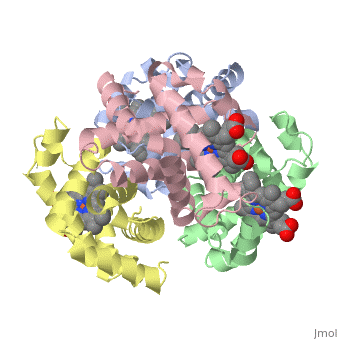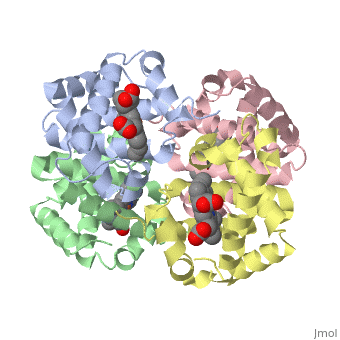
|PDB= 1a3n |SIZE=350|CAPTION= <scene name='initialview01'>1a3n</scene>, resolution 1.8Å | |PDB= 1a3n |SIZE=350|CAPTION= <scene name='initialview01'>1a3n</scene>, resolution 1.8Å | ||
|SITE= | |SITE= | ||
|LIGAND= <scene name='pdbligand=HEM:PROTOPORPHYRIN IX CONTAINING FE'>HEM</scene> | |LIGAND= <scene name='pdbligand=HEM:PROTOPORPHYRIN+IX+CONTAINING+FE'>HEM</scene> | ||
|ACTIVITY= | |ACTIVITY= | ||
|GENE= | |GENE= | ||
|DOMAIN= | |||
|RELATEDENTRY= | |||
|RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1a3n FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1a3n OCA], [http://www.ebi.ac.uk/pdbsum/1a3n PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=1a3n RCSB]</span> | |||
}} | }} | ||
| Line 14: | Line 17: | ||
==Overview== | ==Overview== | ||
The structures of deoxy human haemoglobin and an artificial mutant (Tyralpha42-->His) have been solved at 120 K. While overall agreement between these structures and others in the PDB is very good, certain side chains are found to be shifted, absent from the electron-density map or in different rotamers. Non-crystallographic symmetry (NCS) is very well obeyed in the native protein, but not around the site of the changed residue in the mutant. NCS is also not obeyed by the water molecule invariably found in the alpha-chain haem pocket in room-temperature crystal structures of haemoglobin. At 120 K, this water molecule disappears from one alpha chain in the asymmetric unit but not the other. | The structures of deoxy human haemoglobin and an artificial mutant (Tyralpha42-->His) have been solved at 120 K. While overall agreement between these structures and others in the PDB is very good, certain side chains are found to be shifted, absent from the electron-density map or in different rotamers. Non-crystallographic symmetry (NCS) is very well obeyed in the native protein, but not around the site of the changed residue in the mutant. NCS is also not obeyed by the water molecule invariably found in the alpha-chain haem pocket in room-temperature crystal structures of haemoglobin. At 120 K, this water molecule disappears from one alpha chain in the asymmetric unit but not the other. | ||
==About this Structure== | ==About this Structure== | ||
| Line 27: | Line 27: | ||
[[Category: Tame, J.]] | [[Category: Tame, J.]] | ||
[[Category: Vallone, B.]] | [[Category: Vallone, B.]] | ||
[[Category: erythrocyte]] | [[Category: erythrocyte]] | ||
[[Category: heme]] | [[Category: heme]] | ||
| Line 33: | Line 32: | ||
[[Category: respiratory protein]] | [[Category: respiratory protein]] | ||
''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Sun Mar 30 18:32:31 2008'' | ||
Revision as of 18:32, 30 March 2008
| |||||||
| , resolution 1.8Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | |||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
DEOXY HUMAN HEMOGLOBIN
OverviewOverview
The structures of deoxy human haemoglobin and an artificial mutant (Tyralpha42-->His) have been solved at 120 K. While overall agreement between these structures and others in the PDB is very good, certain side chains are found to be shifted, absent from the electron-density map or in different rotamers. Non-crystallographic symmetry (NCS) is very well obeyed in the native protein, but not around the site of the changed residue in the mutant. NCS is also not obeyed by the water molecule invariably found in the alpha-chain haem pocket in room-temperature crystal structures of haemoglobin. At 120 K, this water molecule disappears from one alpha chain in the asymmetric unit but not the other.
About this StructureAbout this Structure
1A3N is a Protein complex structure of sequences from Homo sapiens. Full crystallographic information is available from OCA.
ReferenceReference
The structures of deoxy human haemoglobin and the mutant Hb Tyralpha42His at 120 K., Tame JR, Vallone B, Acta Crystallogr D Biol Crystallogr. 2000 Jul;56(Pt 7):805-11. PMID:10930827
Page seeded by OCA on Sun Mar 30 18:32:31 2008