Single stranded binding protein: Difference between revisions
No edit summary |
No edit summary |
||
| Line 9: | Line 9: | ||
SSB proteins have been identified in many different organisms, but the most well understood SSB remains the SSB of E. coli. E. coli SSB is a homotetramer consisting of four identical subunits which are each about 19 kDa in size <ref>PMID:2087220</ref>. There are two different binding modes of the E. coli SSB when it complexes with ssDNA (Lohman). Regulation of these modes has been found to be dependent on salt concentration, in addition to other unknown factors. Under low salt conditions, the protein is less efficient as only two of the four identical subunits of E. coli SSB were found to bind to the ssDNA <ref>PMID:11993998</ref>. Under high salt concentrations, however, all four subunits of the homotetramer bind to the ssDNA, increasing the number of nucleotides in contact with the SSB and thus favoring SSB-ssDNA interactions. Depending on the salt concentration and other factors, estimates of the size of the site of interaction between SSB and ssDNA range anywhere from 30 to 73 nucleotides for each tetramer <ref>PMID:11993998</ref>. | SSB proteins have been identified in many different organisms, but the most well understood SSB remains the SSB of E. coli. E. coli SSB is a homotetramer consisting of four identical subunits which are each about 19 kDa in size <ref>PMID:2087220</ref>. There are two different binding modes of the E. coli SSB when it complexes with ssDNA (Lohman). Regulation of these modes has been found to be dependent on salt concentration, in addition to other unknown factors. Under low salt conditions, the protein is less efficient as only two of the four identical subunits of E. coli SSB were found to bind to the ssDNA <ref>PMID:11993998</ref>. Under high salt concentrations, however, all four subunits of the homotetramer bind to the ssDNA, increasing the number of nucleotides in contact with the SSB and thus favoring SSB-ssDNA interactions. Depending on the salt concentration and other factors, estimates of the size of the site of interaction between SSB and ssDNA range anywhere from 30 to 73 nucleotides for each tetramer <ref>PMID:11993998</ref>. | ||
Active E. coli SSB is made of a homotetramer with extensive DNA binding domains that bind to a single strand of DNA (PMID: 2087220). The tetramers consist of α-helices, β-sheets, and random coils. Each subunit contains an α-helix and several β-sheets. The secondary structure also includes a NH2 terminus, which consists of multiple positively charged amino acids. The DNA-binding domain lies within 115 amino acid residues from this terminus. The COOH terminus includes many acidic amino acids | Active E. coli SSB is made of a homotetramer with extensive DNA binding domains that bind to a single strand of DNA (PMID: 2087220). The tetramers consist of α-helices, β-sheets, and random coils. Each subunit contains an α-helix and several β-sheets. The secondary structure also includes a NH2 terminus, which consists of multiple positively charged amino acids. The DNA-binding domain lies within 115 amino acid residues from this terminus. The COOH terminus includes many acidic amino acids <ref>PMID: 2087220</ref>. | ||
</StructureSection> | </StructureSection> | ||
== | ==Binding Interactions in the Active Site== | ||
<StructureSection load='2vw9' size='400' side='left' frame='true' caption='Structure of Single Stranded DNA-Binding Protein from Helicobacter | Single-stranded DNA can interact with SSB through hydrogen bonds, stacking, or electronegative interactions. Though SSB proteins are found in a variety of different organisms, most interactions between SSB and ssDNA happen through the common structural motif of an oligosaccharide/oligonucleotide binding site, referred to as the OB fold <ref>Shamoo, Yousif. “Single Stranded DNA binding proteins.” ‘’Encyclopedia of Life Sciences.’’ MacMillan Publishers Ltd, Nature Publishing Group; 2002</ref>. The OB fold allows SSB to bind preferentially to ssDNA. Each subunit of a SSB has an OB fold (the SSB of E. coli thus has four OB folds, one per each of its four identical subunits). This fold consists of a 5-stranded β barrel that ends in an α-helix. | ||
Several specific amino acid residues play essential roles in the binding of ssDNA to SSB. PHE60 is a key residue involved in binding the ssDNA to the protein, as it has been shown to be the site for cross-linking. Tryptophan and Lysine residues are important in binding as well, as evidenced by modification treatments of lysine and tryptophan residues resulting a complete loss of binding activity for the protein. The two tryptophan residues involved in ssDNA binding are Trp40 and Trp54, which were determined by mutagenesis <ref>PMID:2087220</ref>. | |||
One more key residue in the binding site, His55, was determined by site-specific mutagenesis, as when His55 is substituted with Leu it decreases the overall binding affinity for ssDNA. All of these residues are found in a hydrophobic region, which is suitable for nucleotide base interactions. Treatments that modified arginine, cysteine, or tyrosine residues had no effect on binding of SSB to DNA, suggesting that these amino acids are not involved in significant interactions of the protein with the ssDNA. | |||
==Interactions Between E. coli SSB and other Proteins== | |||
Most of the molecule loses flexibility after ssDNA binding. However, three phenylalanine residues (Phe147, Phe171, Phe177) in the COOH terminal domain remain flexible, even after DNA binding, suggesting that the COOH terminus has something to do with protein binding (PMID: 2087220). An experiment where Phe177 was changed to Cys resulted in a protein that could not replicate DNA. This replication defect stemming from the lost phenylalanine residue was likely a result of the inability of the altered C-terminal region to bind other proteins necessary for replication (PMID:2453719). | |||
Gly15 is believed to play an important role in binding the RecA protein. Mutations in Gly15 have been shown to have extreme effects on recombinational repair. SSB is also thought to bind with exonuclease I, DNA polymerase II, and a protein n, which is a part of the primosome complex and used to help synthesize RNA primers for the lagging strand <ref>PMID: 2087220</ref>. | |||
==Other SSB Structures== | |||
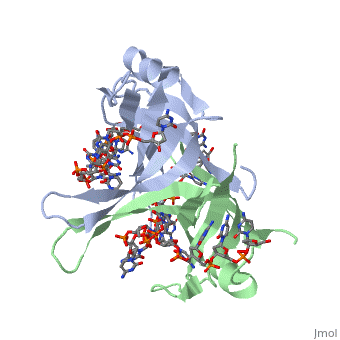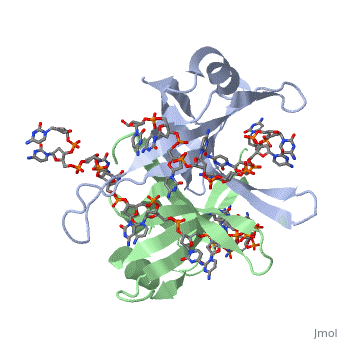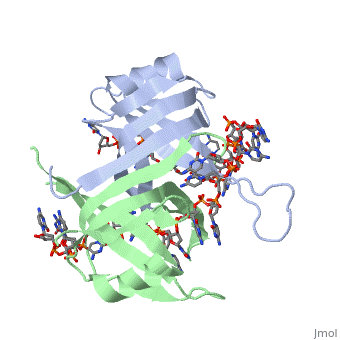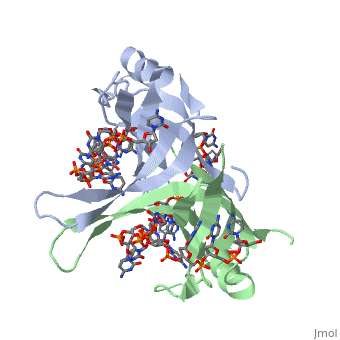
<StructureSection load='2vw9' size='400' side='left' frame='true' caption='Structure of Single Stranded DNA-Binding Protein from ''Helicobacter pylori' 'bound to ssDNA (PDB entry [[2vw9]])' scene=''> Though the SSB of E. coli is perhaps the best characterized, ssDNA binding proteins of many other organisms have also been identified. Some proteins, such as the SSB of E. coli and human mitochondrial SSBs, bind as tetramers to ssDNA. However, the SSB can have from one to as many as four OB-fold containing subunits in its structure. For example, the structure of the SSB from Helicobacter pylori shown to the left is a homodimer composed of two identical subunits, each with the an OB-fold motif made of similar secondary structure elements such as an α-helix and several β-sheets. Just like in the E. coli SSB, the OB-fold area in each subunit is used for single-stranded nucleic acid binding. A phenylalanine residue (PHE56) is again integral to ssDNA binding, as it is a site of cross-linking. Tryptophan and lysine residues again play an important role in binding of ssDNA to the protein. | |||
As single-stranded DNA binding proteins are utilized in some of the most important aspects of DNA metabolism, they are used extensively in DNA replication, repair and recombination. Most SSBs use one or more subunits with an OB-fold motif to bind securely and preferentially to ssDNA. A few specific SSBs (such as RecA and adenovirus DBP) do not use the OB-fold, instead relying on electrostatic and stacking interactions as well as hydrogen bonding. | |||
</StructureSection> | |||
====Binding Interactions in the Active Site==== | ====Binding Interactions in the Active Site==== | ||
| Line 35: | Line 49: | ||
SSB has also been thought to bind with exonuclease I, DNA polymerase II, | SSB has also been thought to bind with exonuclease I, DNA polymerase II, | ||
and a protein n, which is used to help synthesize RNA primers for the lagging strand. SSB can also help regulate transcription by competing with other proteins for binding spaces on DNA. SSB has a higher affinity for DNA than most other proteins, and those proteins are not able to remove SSB from DNA and bind themselves. This type of mechanism can not only regulate transcription, but it can provide protection for the DNA <ref>PMID: 2087220</ref>. | and a protein n, which is used to help synthesize RNA primers for the lagging strand. SSB can also help regulate transcription by competing with other proteins for binding spaces on DNA. SSB has a higher affinity for DNA than most other proteins, and those proteins are not able to remove SSB from DNA and bind themselves. This type of mechanism can not only regulate transcription, but it can provide protection for the DNA <ref>PMID: 2087220</ref>. | ||
</StructureSection> | </StructureSection> | ||
Revision as of 02:38, 3 November 2013
Sandbox Single Stranded DNA-Binding Protein (SSB)
OverviewOverview
Single-stranded DNA-binding protein, or SSB binds to single-stranded regions of DNA. This binding serves a variety of functions - it prevents the strands from hardening too early during replication, it protects the single-stranded DNA from being broken down by nucleases, and it removes the secondary structure of the strands so that other enzymes are able to access them and act effectively upon the strands[1].
Single-stranded DNA (ssDNA) is utilized primarily during the course of major aspects of DNA metabolism such as replication, recombination and repair (PMID: 2087220). In addition to stabilizing ssDNA, SSB proteins also bind to and control the function of many other proteins that are involved in all of three of these major DNA metabolic processes. During DNA replication, SSB molecules bind to the newly separated individual DNA strands, keeping the strands separated by holding them in place so that each strand can serve as a template for new DNA synthesis[2].
Structure of E. coli SSBStructure of E. coli SSB
SSB proteins have been identified in many different organisms, but the most well understood SSB remains the SSB of E. coli. E. coli SSB is a homotetramer consisting of four identical subunits which are each about 19 kDa in size [3]. There are two different binding modes of the E. coli SSB when it complexes with ssDNA (Lohman). Regulation of these modes has been found to be dependent on salt concentration, in addition to other unknown factors. Under low salt conditions, the protein is less efficient as only two of the four identical subunits of E. coli SSB were found to bind to the ssDNA [4]. Under high salt concentrations, however, all four subunits of the homotetramer bind to the ssDNA, increasing the number of nucleotides in contact with the SSB and thus favoring SSB-ssDNA interactions. Depending on the salt concentration and other factors, estimates of the size of the site of interaction between SSB and ssDNA range anywhere from 30 to 73 nucleotides for each tetramer [5]. Active E. coli SSB is made of a homotetramer with extensive DNA binding domains that bind to a single strand of DNA (PMID: 2087220). The tetramers consist of α-helices, β-sheets, and random coils. Each subunit contains an α-helix and several β-sheets. The secondary structure also includes a NH2 terminus, which consists of multiple positively charged amino acids. The DNA-binding domain lies within 115 amino acid residues from this terminus. The COOH terminus includes many acidic amino acids [6]. |
| ||||||||||
Binding Interactions in the Active SiteBinding Interactions in the Active Site
Single-stranded DNA can interact with SSB through hydrogen bonds, stacking, or electronegative interactions. Though SSB proteins are found in a variety of different organisms, most interactions between SSB and ssDNA happen through the common structural motif of an oligosaccharide/oligonucleotide binding site, referred to as the OB fold [7]. The OB fold allows SSB to bind preferentially to ssDNA. Each subunit of a SSB has an OB fold (the SSB of E. coli thus has four OB folds, one per each of its four identical subunits). This fold consists of a 5-stranded β barrel that ends in an α-helix.
Several specific amino acid residues play essential roles in the binding of ssDNA to SSB. PHE60 is a key residue involved in binding the ssDNA to the protein, as it has been shown to be the site for cross-linking. Tryptophan and Lysine residues are important in binding as well, as evidenced by modification treatments of lysine and tryptophan residues resulting a complete loss of binding activity for the protein. The two tryptophan residues involved in ssDNA binding are Trp40 and Trp54, which were determined by mutagenesis [8].
One more key residue in the binding site, His55, was determined by site-specific mutagenesis, as when His55 is substituted with Leu it decreases the overall binding affinity for ssDNA. All of these residues are found in a hydrophobic region, which is suitable for nucleotide base interactions. Treatments that modified arginine, cysteine, or tyrosine residues had no effect on binding of SSB to DNA, suggesting that these amino acids are not involved in significant interactions of the protein with the ssDNA.
Interactions Between E. coli SSB and other ProteinsInteractions Between E. coli SSB and other Proteins
Most of the molecule loses flexibility after ssDNA binding. However, three phenylalanine residues (Phe147, Phe171, Phe177) in the COOH terminal domain remain flexible, even after DNA binding, suggesting that the COOH terminus has something to do with protein binding (PMID: 2087220). An experiment where Phe177 was changed to Cys resulted in a protein that could not replicate DNA. This replication defect stemming from the lost phenylalanine residue was likely a result of the inability of the altered C-terminal region to bind other proteins necessary for replication (PMID:2453719). Gly15 is believed to play an important role in binding the RecA protein. Mutations in Gly15 have been shown to have extreme effects on recombinational repair. SSB is also thought to bind with exonuclease I, DNA polymerase II, and a protein n, which is a part of the primosome complex and used to help synthesize RNA primers for the lagging strand [9].
Other SSB StructuresOther SSB Structures
| Though the SSB of E. coli is perhaps the best characterized, ssDNA binding proteins of many other organisms have also been identified. Some proteins, such as the SSB of E. coli and human mitochondrial SSBs, bind as tetramers to ssDNA. However, the SSB can have from one to as many as four OB-fold containing subunits in its structure. For example, the structure of the SSB from Helicobacter pylori shown to the left is a homodimer composed of two identical subunits, each with the an OB-fold motif made of similar secondary structure elements such as an α-helix and several β-sheets. Just like in the E. coli SSB, the OB-fold area in each subunit is used for single-stranded nucleic acid binding. A phenylalanine residue (PHE56) is again integral to ssDNA binding, as it is a site of cross-linking. Tryptophan and lysine residues again play an important role in binding of ssDNA to the protein. As single-stranded DNA binding proteins are utilized in some of the most important aspects of DNA metabolism, they are used extensively in DNA replication, repair and recombination. Most SSBs use one or more subunits with an OB-fold motif to bind securely and preferentially to ssDNA. A few specific SSBs (such as RecA and adenovirus DBP) do not use the OB-fold, instead relying on electrostatic and stacking interactions as well as hydrogen bonding. | ||||||||||
Binding Interactions in the Active SiteBinding Interactions in the Active Site
ssDNA can interact with binding proteins through hydrogen bonds, stacking, or electronegative interactions. Most interactions between SSB and ssDNA happen through the OB fold. OB stands for oligosaccharide/oligonucleotide binding site. This fold consists of a 5-stranded β barrel that ends in an α-helix. is an important DNA binding site. It has been shown to be the site for cross-linking. Tryptophan and Lysine residues are important in binding as well. Treatments resulting in modification of arginine, cysteine, or tyrosine residues had no effect on binding of SSB to DNA, whereas modification of either lysine residues (with acetic anhydride) or tryptophan residues (with N-bromosuccinimide) led to complete loss of binding activity [10].
SSB-Protein InteractionsSSB-Protein Interactions
SSB can form complexes with many other proteins. This trait can keep enzymes needed for damage repair, transcription, etc. near the ssDNA and it is thought that SSB can even help to stimulate these enzymes to carry out their jobs. When DNA binds SSB, most of the molecule loses flexibility. But the COOH terminal domain remain flexible, even after DNA binding. It is believed that the COOH terminus has something to do with protein binding [11].
SSB will interact with the protein RecA to enable recombination, because RecA will recognize SSB and replace it on the strand. In DNA repair, SSB will bind to the damaged strand to protect it. And eventually it will attract repair enzymes which will replace SSB and begin repair mechanisms.
SSB has also been thought to bind with exonuclease I, DNA polymerase II, and a protein n, which is used to help synthesize RNA primers for the lagging strand. SSB can also help regulate transcription by competing with other proteins for binding spaces on DNA. SSB has a higher affinity for DNA than most other proteins, and those proteins are not able to remove SSB from DNA and bind themselves. This type of mechanism can not only regulate transcription, but it can provide protection for the DNA [12]. </StructureSection>
See AlsoSee Also
ReferencesReferences
- ↑ PMID: 2087220)
- ↑ Berg JM, Tymoczko JL, Stryer L. Biochemistry. 6th edition. New York: W H Freeman; 2006.
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Kozlov AG, Lohman TM. Stopped-flow studies of the kinetics of single-stranded DNA binding and wrapping around the Escherichia coli SSB tetramer. Biochemistry. 2002 May 14;41(19):6032-44. PMID:11993998
- ↑ Kozlov AG, Lohman TM. Stopped-flow studies of the kinetics of single-stranded DNA binding and wrapping around the Escherichia coli SSB tetramer. Biochemistry. 2002 May 14;41(19):6032-44. PMID:11993998
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Shamoo, Yousif. “Single Stranded DNA binding proteins.” ‘’Encyclopedia of Life Sciences.’’ MacMillan Publishers Ltd, Nature Publishing Group; 2002
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220