FhuD: Difference between revisions
No edit summary |
No edit summary |
||
| Line 2: | Line 2: | ||
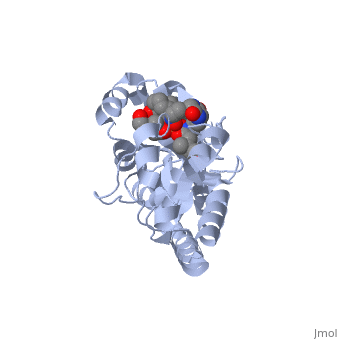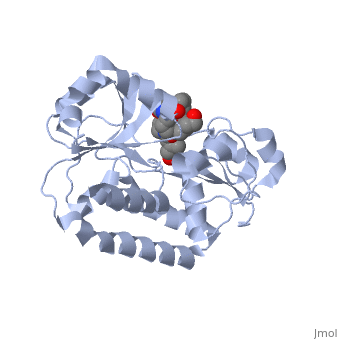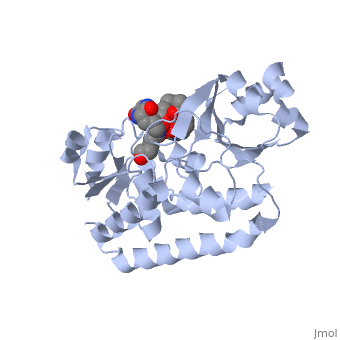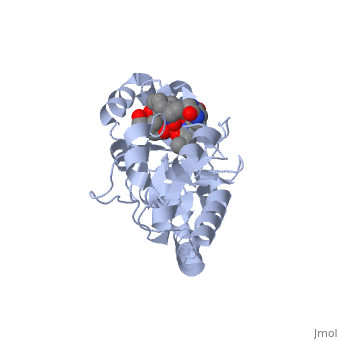
=='''PERIPLASMIC FERRIC SIDEROPHORE BINDING PROTEIN FHUD COMPLEXED WITH COPROGEN (1esz)'''== | =='''PERIPLASMIC FERRIC SIDEROPHORE BINDING PROTEIN FHUD COMPLEXED WITH COPROGEN (1esz)'''== | ||
<applet load='1esz' size='300' frame='true' align='right' caption=' | |||
---- | |||
<applet load='1esz' size='300' frame='true' align='right' caption='STRUCTURE OF THE PERIPLASMIC FERRIC SIDEROPHORE BINDING PROTEIN FHUD COMPLEXED WITH COPROGEN as determined by Clarke ''et al.''' /> | |||
==OVERVIEW== | ==OVERVIEW== | ||
Siderophore-binding proteins can be found in both Gram-positive and Gram-negative bacteria in two divisions: hydroxamates and catecholates. In Escherichia coli. (E. coli) the ATP-binding cassette- type (ABC-type) protein FhuD is a common periplasmic protein which facilitates the transport of a variety of hydoxamate siderophores to the inner membrane-associated proteins FhuB and FhuC. The structure of FhuD is atypical for periplasmic ligand binding protein due to its bilobal mixture of two α/β domains connected by long α-helix. | Siderophore-binding proteins can be found in both Gram-positive and Gram-negative bacteria in two divisions: hydroxamates and catecholates. In Escherichia coli. (''E. coli'') the ATP-binding cassette- type (ABC-type) protein FhuD is a common periplasmic protein which facilitates the transport of a variety of hydoxamate siderophores to the inner membrane-associated proteins FhuB and FhuC. The structure of FhuD is atypical for periplasmic ligand binding protein due to its bilobal mixture of two α/β domains connected by long α-helix. | ||
==PROTEIN STRUCTURE== | ==PROTEIN STRUCTURE== | ||
FhuD is atypical for periplasmic ligand binding proteins. It is a bilobal kidney bean shape containing two domains which are connected by a 23-residue kinked α-helix. The N-terminal domain twisted fived-stranded parallel β-sheet whereas the C-terminal domain has a mixed five stranded β-sheet; both are | FhuD is atypical for periplasmic ligand binding proteins. It is a bilobal kidney bean shape with approximate dimensions 60 Å ´ 30 Å ´ 40 Å.(ref both) containing two domains which are connected by a 23-residue kinked α-helix. The N-terminal domain (residues 27–141) twisted fived-stranded parallel β-sheet with 3-2-1-4-5 linking topology whereas the C-terminal domain (residues 166–288) has a mixed five stranded β-sheet 3-2-1-4-5 linking topology; both are enclosed by α-helices. Between the two domains lies the shallow siderophore binding site approximate 10 Å deep (REF) which forms depression or “pocket.” This pocket is lined with hydrophobic residues which side chain residues are able to create stabilizing hydrogen bond with the accepted siderophore. This is large enough to accommodate the hydrophobic orinthyl linkers of the siderophore. Through rearrangements of the residues of the binding pocket and interactions with the iron-hydroxamate centers of the siderophore, recognition can occur with structurally diverse siderophores. The binding diversity is further increased since the siderophore backbones do not interact with the proteins. | ||
==PROTEIN FUNCTION== | ==PROTEIN FUNCTION== | ||
[[User:Leni Rose|Leni Rose]] 04:57, 13 March 2010 (IST) | [[User:Leni Rose|Leni Rose]] 04:57, 13 March 2010 (IST) | ||
Revision as of 06:42, 13 March 2010
PERIPLASMIC FERRIC SIDEROPHORE BINDING PROTEIN FHUD COMPLEXED WITH COPROGEN (1esz)PERIPLASMIC FERRIC SIDEROPHORE BINDING PROTEIN FHUD COMPLEXED WITH COPROGEN (1esz)
| Please do NOT make changes to this Sandbox until after April 23, 2010. Sandboxes 151-200 are reserved until then for use by the Chemistry 307 class at UNBC taught by Prof. Andrea Gorrell. |
|
OVERVIEWOVERVIEW
Siderophore-binding proteins can be found in both Gram-positive and Gram-negative bacteria in two divisions: hydroxamates and catecholates. In Escherichia coli. (E. coli) the ATP-binding cassette- type (ABC-type) protein FhuD is a common periplasmic protein which facilitates the transport of a variety of hydoxamate siderophores to the inner membrane-associated proteins FhuB and FhuC. The structure of FhuD is atypical for periplasmic ligand binding protein due to its bilobal mixture of two α/β domains connected by long α-helix.
PROTEIN STRUCTUREPROTEIN STRUCTURE
FhuD is atypical for periplasmic ligand binding proteins. It is a bilobal kidney bean shape with approximate dimensions 60 Å ´ 30 Å ´ 40 Å.(ref both) containing two domains which are connected by a 23-residue kinked α-helix. The N-terminal domain (residues 27–141) twisted fived-stranded parallel β-sheet with 3-2-1-4-5 linking topology whereas the C-terminal domain (residues 166–288) has a mixed five stranded β-sheet 3-2-1-4-5 linking topology; both are enclosed by α-helices. Between the two domains lies the shallow siderophore binding site approximate 10 Å deep (REF) which forms depression or “pocket.” This pocket is lined with hydrophobic residues which side chain residues are able to create stabilizing hydrogen bond with the accepted siderophore. This is large enough to accommodate the hydrophobic orinthyl linkers of the siderophore. Through rearrangements of the residues of the binding pocket and interactions with the iron-hydroxamate centers of the siderophore, recognition can occur with structurally diverse siderophores. The binding diversity is further increased since the siderophore backbones do not interact with the proteins.
PROTEIN FUNCTIONPROTEIN FUNCTION
Leni Rose 04:57, 13 March 2010 (IST)