2abx: Difference between revisions
No edit summary |
No edit summary |
||
| (12 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
< | ==THE CRYSTAL STRUCTURE OF ALPHA-BUNGAROTOXIN AT 2.5 ANGSTROMS RESOLUTION. RELATION TO SOLUTION STRUCTURE AND BINDING TO ACETYLCHOLINE RECEPTOR== | ||
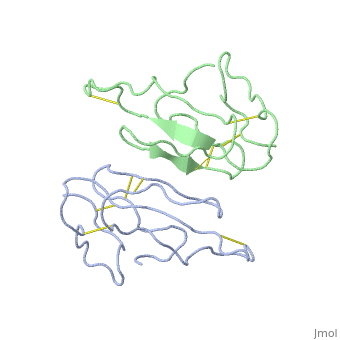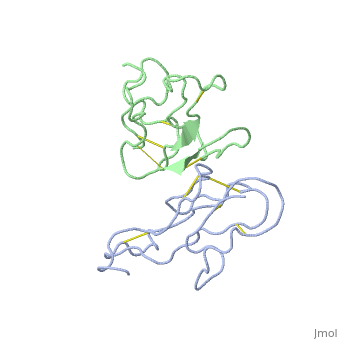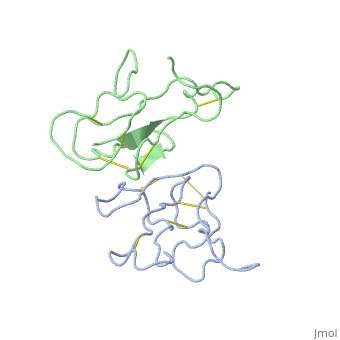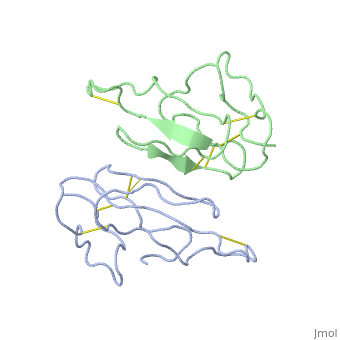
<StructureSection load='2abx' size='340' side='right'caption='[[2abx]], [[Resolution|resolution]] 2.50Å' scene=''> | |||
You may | == Structural highlights == | ||
<table><tr><td colspan='2'>[[2abx]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Bungarus_multicinctus Bungarus multicinctus]. This structure supersedes the now removed PDB entry [http://oca.weizmann.ac.il/oca-bin/send-pdb?obs=1&id=1abx 1abx]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2ABX OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2ABX FirstGlance]. <br> | |||
or | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.5Å</td></tr> | ||
-- | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2abx FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2abx OCA], [https://pdbe.org/2abx PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2abx RCSB], [https://www.ebi.ac.uk/pdbsum/2abx PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2abx ProSAT]</span></td></tr> | ||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/3L21A_BUNMU 3L21A_BUNMU] Binds with high affinity to muscular (tested on Torpedo marmorata, Kd=0.4 nM) and neuronal (tested on chimeric alpha-7/CHRNA7, Kd=0.95 nM) nicotinic acetylcholine receptor (nAChR) and inhibits acetylcholine from binding to the receptor, thereby impairing neuromuscular and neuronal transmission (PubMed:9305882). It also shows an activity on GABA(A) receptors (PubMed:16549768, PubMed:25634239). It antagonises GABA-activated currents with high potency when tested on primary hippocampal neurons (PubMed:25634239). It inhibits recombinantly expressed GABA(A) receptors composed of alpha-2-beta-2-gamma-2 (GABRA2-GABRB2-GABRG2) subunits with high potency (62.3% inhibition at 20 uM of toxin) (PubMed:25634239). It also shows a weaker inhibition on GABA(A) receptors composed of alpha-1-beta-2-gamma-2 (GABRA1-GABRB2-GABRG2) subunits, alpha-4-beta-2-gamma-2 (GABRA4-GABRB2-GABRG2) subunits, and alpha-5-beta-2-gamma-2 (GABRA5-GABRB2-GABRG2) subunits (PubMed:25634239). A very weak inhibition is also observed on GABA(A) receptor composed of alpha-1-beta-3-gamma-2 (GABRA1-GABRB3-GABRG2) (PubMed:26221036). It has also been shown to bind and inhibit recombinant GABA(A) receptor beta-3/GABRB3 subunit (Kd=about 50 nM) (PubMed:16549768). In addition, it blocks the extracellular increase of dopamine evoked by nicotine only at the higher dose (4.2 uM) (PubMed:9840221).<ref>PMID:16549768</ref> <ref>PMID:25634239</ref> <ref>PMID:9305882</ref> <ref>PMID:9840221</ref> | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/ab/2abx_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=2abx ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
We report collection of 2.5 A resolution X-ray diffraction data from newly grown crystals of the rare 'small unit cell' form of the long snake neurotoxin, alpha-bungarotoxin. The previous model of the molecule has been rebuilt, and refined using least-square methods to a crystallographic residual of 0.24 at 2.5 A resolution. alpha-Bungarotoxin's crystal structure is compared with the crystal structures of two other snake neurotoxins (cobratoxin and erabutoxin), and with its solution structure inferred from spectroscopic studies. Significant differences include less beta-sheet in bungarotoxin's crystal structure than in solution, or in the crystal structures of other neurotoxins, and an unusual orientation in the crystal of the invariant tryptophan. The functional, binding surface of bungarotoxin is described; it consists primarily of hydrophobic and hydrogen-bonding groups and only a few charged side-chains. The structure is compared with experimental binding parameters for neurotoxins. | |||
The crystal structure of alpha-bungarotoxin at 2.5 A resolution: relation to solution structure and binding to acetylcholine receptor.,Love RA, Stroud RM Protein Eng. 1986 Oct-Nov;1(1):37-46. PMID:3507686<ref>PMID:3507686</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 2abx" style="background-color:#fffaf0;"></div> | |||
==See Also== | |||
*[[Bungarotoxin 3D structures|Bungarotoxin 3D structures]] | |||
== References == | |||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
== | |||
== | |||
< | |||
[[Category: Bungarus multicinctus]] | [[Category: Bungarus multicinctus]] | ||
[[Category: | [[Category: Large Structures]] | ||
[[Category: | [[Category: Love R]] | ||
[[Category: | [[Category: Stroud R]] | ||
Latest revision as of 11:16, 15 May 2024
THE CRYSTAL STRUCTURE OF ALPHA-BUNGAROTOXIN AT 2.5 ANGSTROMS RESOLUTION. RELATION TO SOLUTION STRUCTURE AND BINDING TO ACETYLCHOLINE RECEPTORTHE CRYSTAL STRUCTURE OF ALPHA-BUNGAROTOXIN AT 2.5 ANGSTROMS RESOLUTION. RELATION TO SOLUTION STRUCTURE AND BINDING TO ACETYLCHOLINE RECEPTOR
Structural highlights
Function3L21A_BUNMU Binds with high affinity to muscular (tested on Torpedo marmorata, Kd=0.4 nM) and neuronal (tested on chimeric alpha-7/CHRNA7, Kd=0.95 nM) nicotinic acetylcholine receptor (nAChR) and inhibits acetylcholine from binding to the receptor, thereby impairing neuromuscular and neuronal transmission (PubMed:9305882). It also shows an activity on GABA(A) receptors (PubMed:16549768, PubMed:25634239). It antagonises GABA-activated currents with high potency when tested on primary hippocampal neurons (PubMed:25634239). It inhibits recombinantly expressed GABA(A) receptors composed of alpha-2-beta-2-gamma-2 (GABRA2-GABRB2-GABRG2) subunits with high potency (62.3% inhibition at 20 uM of toxin) (PubMed:25634239). It also shows a weaker inhibition on GABA(A) receptors composed of alpha-1-beta-2-gamma-2 (GABRA1-GABRB2-GABRG2) subunits, alpha-4-beta-2-gamma-2 (GABRA4-GABRB2-GABRG2) subunits, and alpha-5-beta-2-gamma-2 (GABRA5-GABRB2-GABRG2) subunits (PubMed:25634239). A very weak inhibition is also observed on GABA(A) receptor composed of alpha-1-beta-3-gamma-2 (GABRA1-GABRB3-GABRG2) (PubMed:26221036). It has also been shown to bind and inhibit recombinant GABA(A) receptor beta-3/GABRB3 subunit (Kd=about 50 nM) (PubMed:16549768). In addition, it blocks the extracellular increase of dopamine evoked by nicotine only at the higher dose (4.2 uM) (PubMed:9840221).[1] [2] [3] [4] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedWe report collection of 2.5 A resolution X-ray diffraction data from newly grown crystals of the rare 'small unit cell' form of the long snake neurotoxin, alpha-bungarotoxin. The previous model of the molecule has been rebuilt, and refined using least-square methods to a crystallographic residual of 0.24 at 2.5 A resolution. alpha-Bungarotoxin's crystal structure is compared with the crystal structures of two other snake neurotoxins (cobratoxin and erabutoxin), and with its solution structure inferred from spectroscopic studies. Significant differences include less beta-sheet in bungarotoxin's crystal structure than in solution, or in the crystal structures of other neurotoxins, and an unusual orientation in the crystal of the invariant tryptophan. The functional, binding surface of bungarotoxin is described; it consists primarily of hydrophobic and hydrogen-bonding groups and only a few charged side-chains. The structure is compared with experimental binding parameters for neurotoxins. The crystal structure of alpha-bungarotoxin at 2.5 A resolution: relation to solution structure and binding to acetylcholine receptor.,Love RA, Stroud RM Protein Eng. 1986 Oct-Nov;1(1):37-46. PMID:3507686[5] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||