1ppb: Difference between revisions
New page: left|200px<br /> <applet load="1ppb" size="450" color="white" frame="true" align="right" spinBox="true" caption="1ppb, resolution 1.92Å" /> '''THE REFINED 1.9 ANG... |
No edit summary |
||
| (21 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
== | ==THE REFINED 1.9 ANGSTROMS CRYSTAL STRUCTURE OF HUMAN ALPHA-THROMBIN: INTERACTION WITH D-PHE-PRO-ARG CHLOROMETHYLKETONE AND SIGNIFICANCE OF THE TYR-PRO-PRO-TRP INSERTION SEGMENT== | ||
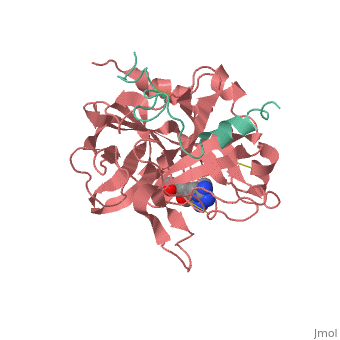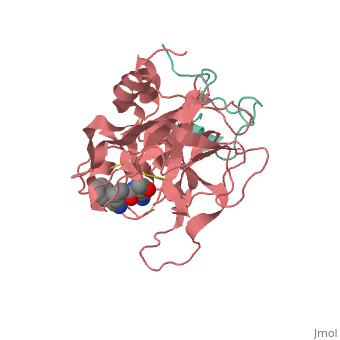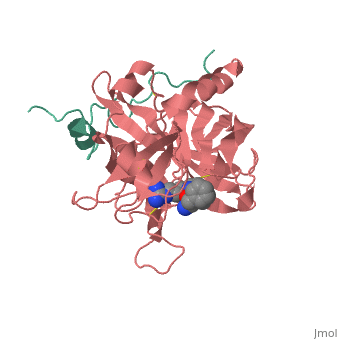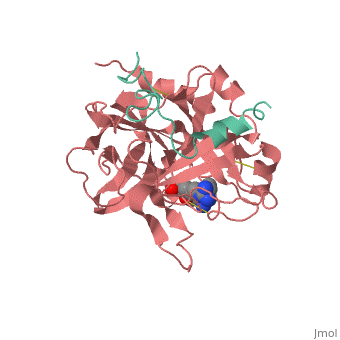
A stoichiometric complex formed between human alpha-thrombin and | <StructureSection load='1ppb' size='340' side='right'caption='[[1ppb]], [[Resolution|resolution]] 1.92Å' scene=''> | ||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[1ppb]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. The January 2002 RCSB PDB [https://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''Thrombin'' by David S. Goodsell is [https://dx.doi.org/10.2210/rcsb_pdb/mom_2002_1 10.2210/rcsb_pdb/mom_2002_1]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1PPB OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1PPB FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.92Å</td></tr> | |||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=0G6:D-PHENYLALANYL-N-[(2S,3S)-6-{[AMINO(IMINIO)METHYL]AMINO}-1-CHLORO-2-HYDROXYHEXAN-3-YL]-L-PROLINAMIDE'>0G6</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1ppb FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1ppb OCA], [https://pdbe.org/1ppb PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1ppb RCSB], [https://www.ebi.ac.uk/pdbsum/1ppb PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1ppb ProSAT]</span></td></tr> | |||
</table> | |||
== Disease == | |||
[https://www.uniprot.org/uniprot/THRB_HUMAN THRB_HUMAN] Defects in F2 are the cause of factor II deficiency (FA2D) [MIM:[https://omim.org/entry/613679 613679]. It is a very rare blood coagulation disorder characterized by mucocutaneous bleeding symptoms. The severity of the bleeding manifestations correlates with blood factor II levels.<ref>PMID:14962227</ref> <ref>PMID:6405779</ref> <ref>PMID:3771562</ref> <ref>PMID:3567158</ref> <ref>PMID:3801671</ref> <ref>PMID:3242619</ref> <ref>PMID:2719946</ref> <ref>PMID:1354985</ref> <ref>PMID:1421398</ref> <ref>PMID:1349838</ref> <ref>PMID:7865694</ref> <ref>PMID:7792730</ref> Genetic variations in F2 may be a cause of susceptibility to ischemic stroke (ISCHSTR) [MIM:[https://omim.org/entry/601367 601367]; also known as cerebrovascular accident or cerebral infarction. A stroke is an acute neurologic event leading to death of neural tissue of the brain and resulting in loss of motor, sensory and/or cognitive function. Ischemic strokes, resulting from vascular occlusion, is considered to be a highly complex disease consisting of a group of heterogeneous disorders with multiple genetic and environmental risk factors.<ref>PMID:15534175</ref> Defects in F2 are the cause of thrombophilia due to thrombin defect (THPH1) [MIM:[https://omim.org/entry/188050 188050]. It is a multifactorial disorder of hemostasis characterized by abnormal platelet aggregation in response to various agents and recurrent thrombi formation. Note=A common genetic variation in the 3-prime untranslated region of the prothrombin gene is associated with elevated plasma prothrombin levels and an increased risk of venous thrombosis. Defects in F2 are associated with susceptibility to pregnancy loss, recurrent, type 2 (RPRGL2) [MIM:[https://omim.org/entry/614390 614390]. A common complication of pregnancy, resulting in spontaneous abortion before the fetus has reached viability. The term includes all miscarriages from the time of conception until 24 weeks of gestation. Recurrent pregnancy loss is defined as 3 or more consecutive spontaneous abortions.<ref>PMID:11506076</ref> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/THRB_HUMAN THRB_HUMAN] Thrombin, which cleaves bonds after Arg and Lys, converts fibrinogen to fibrin and activates factors V, VII, VIII, XIII, and, in complex with thrombomodulin, protein C. Functions in blood homeostasis, inflammation and wound healing.<ref>PMID:2856554</ref> | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/pp/1ppb_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1ppb ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
A stoichiometric complex formed between human alpha-thrombin and D-Phe-Pro-Arg chloromethylketone was crystallized in an orthorhombic crystal form. Orientation and position of a starting model derived from homologous modelling were determined by Patterson search methods. The thrombin model was completed in a cyclic modelling-crystallographic refinement procedure to a final R-value of 0.171 for X-ray data to 1.92 A. The structure is in full agreement with published cDNA sequence data. The A-chain, ordered only in its central part, is positioned along the molecular surface opposite to the active site. The B-chain exhibits the characteristic polypeptide fold of trypsin-like proteinases. Several extended insertions form, however, large protuberances; most important for interaction with macromolecular substrates is the characteristic thrombin loop around Tyr60A-Pro60B-Pro60C-Trp60D (chymotrypsinogen numbering) and the enlarged loop around the unique Trp148. The former considerably restricts the active site cleft and seems likely to be responsible for poor binding of most natural proteinase inhibitors to thrombin. The exceptional specificity of D-Phe-Pro-Arg chloromethylketone can be explained by a hydrophobic cage formed by Ile174, Trp215, Leu99, His57, Tyr60A and Trp60D. The narrow active site cleft, with a more polar base and hydrophobic rims, extends towards the arginine-rich surface of loop Lys70-Glu80 that probably represents part of the anionic binding region for hirudin and fibrinogen. | |||
The refined 1.9 A crystal structure of human alpha-thrombin: interaction with D-Phe-Pro-Arg chloromethylketone and significance of the Tyr-Pro-Pro-Trp insertion segment.,Bode W, Mayr I, Baumann U, Huber R, Stone SR, Hofsteenge J EMBO J. 1989 Nov;8(11):3467-75. PMID:2583108<ref>PMID:2583108</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 1ppb" style="background-color:#fffaf0;"></div> | |||
== | ==See Also== | ||
*[[Thrombin|Thrombin]] | |||
*[[Thrombin 3D Structures|Thrombin 3D Structures]] | |||
== References == | |||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
[[Category: | [[Category: Large Structures]] | ||
[[Category: RCSB PDB Molecule of the Month]] | |||
[[Category: Thrombin]] | [[Category: Thrombin]] | ||
[[Category: Bode | [[Category: Bode W]] | ||
Latest revision as of 03:22, 21 November 2024
THE REFINED 1.9 ANGSTROMS CRYSTAL STRUCTURE OF HUMAN ALPHA-THROMBIN: INTERACTION WITH D-PHE-PRO-ARG CHLOROMETHYLKETONE AND SIGNIFICANCE OF THE TYR-PRO-PRO-TRP INSERTION SEGMENTTHE REFINED 1.9 ANGSTROMS CRYSTAL STRUCTURE OF HUMAN ALPHA-THROMBIN: INTERACTION WITH D-PHE-PRO-ARG CHLOROMETHYLKETONE AND SIGNIFICANCE OF THE TYR-PRO-PRO-TRP INSERTION SEGMENT
Structural highlights
DiseaseTHRB_HUMAN Defects in F2 are the cause of factor II deficiency (FA2D) [MIM:613679. It is a very rare blood coagulation disorder characterized by mucocutaneous bleeding symptoms. The severity of the bleeding manifestations correlates with blood factor II levels.[1] [2] [3] [4] [5] [6] [7] [8] [9] [10] [11] [12] Genetic variations in F2 may be a cause of susceptibility to ischemic stroke (ISCHSTR) [MIM:601367; also known as cerebrovascular accident or cerebral infarction. A stroke is an acute neurologic event leading to death of neural tissue of the brain and resulting in loss of motor, sensory and/or cognitive function. Ischemic strokes, resulting from vascular occlusion, is considered to be a highly complex disease consisting of a group of heterogeneous disorders with multiple genetic and environmental risk factors.[13] Defects in F2 are the cause of thrombophilia due to thrombin defect (THPH1) [MIM:188050. It is a multifactorial disorder of hemostasis characterized by abnormal platelet aggregation in response to various agents and recurrent thrombi formation. Note=A common genetic variation in the 3-prime untranslated region of the prothrombin gene is associated with elevated plasma prothrombin levels and an increased risk of venous thrombosis. Defects in F2 are associated with susceptibility to pregnancy loss, recurrent, type 2 (RPRGL2) [MIM:614390. A common complication of pregnancy, resulting in spontaneous abortion before the fetus has reached viability. The term includes all miscarriages from the time of conception until 24 weeks of gestation. Recurrent pregnancy loss is defined as 3 or more consecutive spontaneous abortions.[14] FunctionTHRB_HUMAN Thrombin, which cleaves bonds after Arg and Lys, converts fibrinogen to fibrin and activates factors V, VII, VIII, XIII, and, in complex with thrombomodulin, protein C. Functions in blood homeostasis, inflammation and wound healing.[15] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedA stoichiometric complex formed between human alpha-thrombin and D-Phe-Pro-Arg chloromethylketone was crystallized in an orthorhombic crystal form. Orientation and position of a starting model derived from homologous modelling were determined by Patterson search methods. The thrombin model was completed in a cyclic modelling-crystallographic refinement procedure to a final R-value of 0.171 for X-ray data to 1.92 A. The structure is in full agreement with published cDNA sequence data. The A-chain, ordered only in its central part, is positioned along the molecular surface opposite to the active site. The B-chain exhibits the characteristic polypeptide fold of trypsin-like proteinases. Several extended insertions form, however, large protuberances; most important for interaction with macromolecular substrates is the characteristic thrombin loop around Tyr60A-Pro60B-Pro60C-Trp60D (chymotrypsinogen numbering) and the enlarged loop around the unique Trp148. The former considerably restricts the active site cleft and seems likely to be responsible for poor binding of most natural proteinase inhibitors to thrombin. The exceptional specificity of D-Phe-Pro-Arg chloromethylketone can be explained by a hydrophobic cage formed by Ile174, Trp215, Leu99, His57, Tyr60A and Trp60D. The narrow active site cleft, with a more polar base and hydrophobic rims, extends towards the arginine-rich surface of loop Lys70-Glu80 that probably represents part of the anionic binding region for hirudin and fibrinogen. The refined 1.9 A crystal structure of human alpha-thrombin: interaction with D-Phe-Pro-Arg chloromethylketone and significance of the Tyr-Pro-Pro-Trp insertion segment.,Bode W, Mayr I, Baumann U, Huber R, Stone SR, Hofsteenge J EMBO J. 1989 Nov;8(11):3467-75. PMID:2583108[16] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||