2hh7: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| (14 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
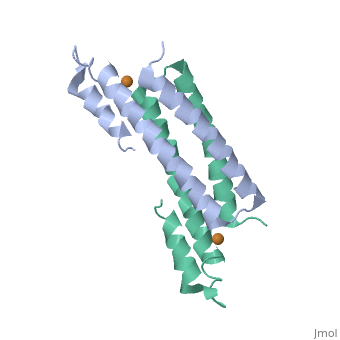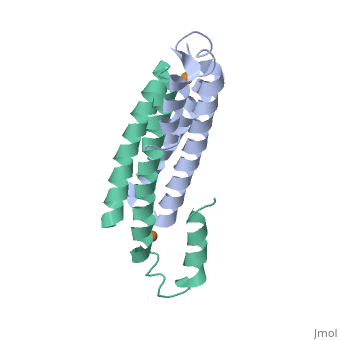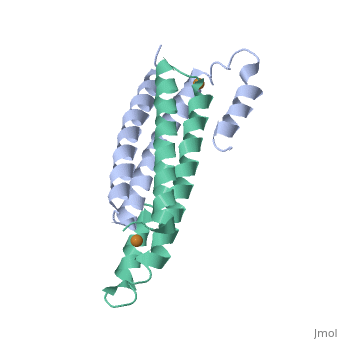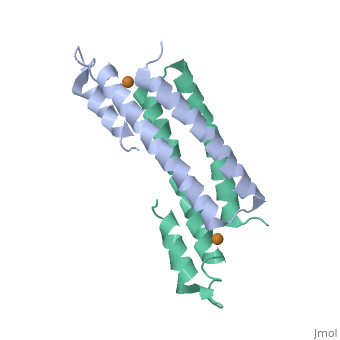
==Crystal Structure of Cu(I) bound CsoR from Mycobacterium tuberculosis.== | |||
<StructureSection load='2hh7' size='340' side='right'caption='[[2hh7]], [[Resolution|resolution]] 2.55Å' scene=''> | |||
| | == Structural highlights == | ||
<table><tr><td colspan='2'>[[2hh7]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Mycobacterium_tuberculosis Mycobacterium tuberculosis]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2HH7 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2HH7 FirstGlance]. <br> | |||
| | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.55Å</td></tr> | ||
| | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=CU1:COPPER+(I)+ION'>CU1</scene></td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2hh7 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2hh7 OCA], [https://pdbe.org/2hh7 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2hh7 RCSB], [https://www.ebi.ac.uk/pdbsum/2hh7 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2hh7 ProSAT]</span></td></tr> | |||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/CSOR_MYCTU CSOR_MYCTU] Copper-sensitive repressor that has a key role in copper homeostasis. It is part of the cso operon involved in the cellular response to increasing concentrations of copper inside the bacterium, which can be highly toxic. In the presence of copper, CsoR fully dissociates from the promoter in the cso operon, leading to the transcription of its genes. Binds to a GC-rich pseudopallindromic sequence, 5'-GTAGCCCACCCCCAGTGGGGTGGGA-3', in the cso promoter region.<ref>PMID:17143269</ref> | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/hh/2hh7_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=2hh7 ConSurf]. | |||
<div style="clear:both"></div> | |||
==See Also== | |||
*[[Copper homeostasis protein|Copper homeostasis protein]] | |||
== References == | |||
== | <references/> | ||
Copper | __TOC__ | ||
</StructureSection> | |||
== | [[Category: Large Structures]] | ||
[[Category: Mycobacterium tuberculosis]] | [[Category: Mycobacterium tuberculosis]] | ||
[[Category: Ramesh A]] | |||
[[Category: Ramesh | [[Category: Sacchettini JC]] | ||
[[Category: Sacchettini | |||
Latest revision as of 12:32, 14 February 2024
Crystal Structure of Cu(I) bound CsoR from Mycobacterium tuberculosis.Crystal Structure of Cu(I) bound CsoR from Mycobacterium tuberculosis.
Structural highlights
FunctionCSOR_MYCTU Copper-sensitive repressor that has a key role in copper homeostasis. It is part of the cso operon involved in the cellular response to increasing concentrations of copper inside the bacterium, which can be highly toxic. In the presence of copper, CsoR fully dissociates from the promoter in the cso operon, leading to the transcription of its genes. Binds to a GC-rich pseudopallindromic sequence, 5'-GTAGCCCACCCCCAGTGGGGTGGGA-3', in the cso promoter region.[1] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. See AlsoReferences
|
| ||||||||||||||||||