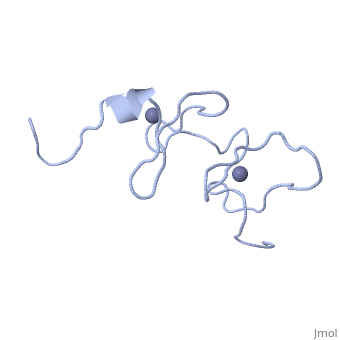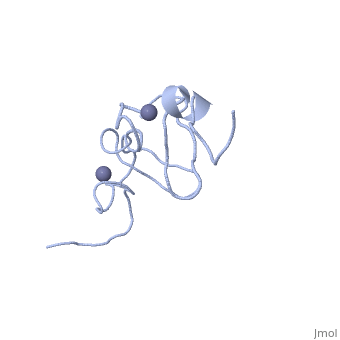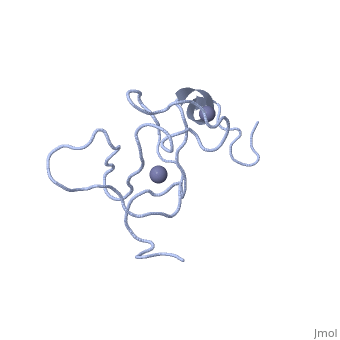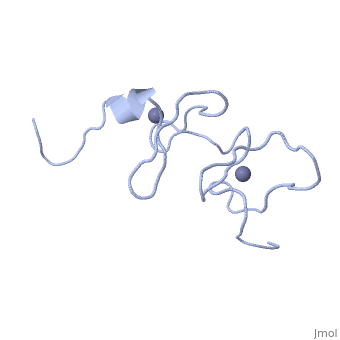1x4l: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| (15 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==Solution structure of LIM domain in Four and a half LIM domains protein 2== | |||
<StructureSection load='1x4l' size='340' side='right'caption='[[1x4l]]' scene=''> | |||
| | == Structural highlights == | ||
| | <table><tr><td colspan='2'>[[1x4l]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full experimental information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1X4L OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1X4L FirstGlance]. <br> | ||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">Solution NMR</td></tr> | |||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=ZN:ZINC+ION'>ZN</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1x4l FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1x4l OCA], [https://pdbe.org/1x4l PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1x4l RCSB], [https://www.ebi.ac.uk/pdbsum/1x4l PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1x4l ProSAT], [https://www.topsan.org/Proteins/RSGI/1x4l TOPSAN]</span></td></tr> | |||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/FHL2_HUMAN FHL2_HUMAN] May function as a molecular transmitter linking various signaling pathways to transcriptional regulation. Negatively regulates the transcriptional repressor E4F1 and may function in cell growth. Inhibits the transcriptional activity of FOXO1 and its apoptotic function by enhancing the interaction of FOXO1 with SIRT1 and FOXO1 deacetylation.<ref>PMID:15692560</ref> <ref>PMID:16652157</ref> <ref>PMID:18853468</ref> | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/x4/1x4l_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1x4l ConSurf]. | |||
<div style="clear:both"></div> | |||
==See Also== | |||
*[[Muscle LIM protein|Muscle LIM protein]] | |||
== References == | |||
== | <references/> | ||
__TOC__ | |||
</StructureSection> | |||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
[[Category: | [[Category: Large Structures]] | ||
[[Category: He | [[Category: He F]] | ||
[[Category: Inoue | [[Category: Inoue M]] | ||
[[Category: Kigawa | [[Category: Kigawa T]] | ||
[[Category: Muto | [[Category: Muto Y]] | ||
[[Category: Shirouzu M]] | |||
[[Category: Shirouzu | [[Category: Terada T]] | ||
[[Category: Terada | [[Category: Yokoyama S]] | ||
[[Category: Yokoyama | |||
Latest revision as of 16:58, 9 May 2024
Solution structure of LIM domain in Four and a half LIM domains protein 2Solution structure of LIM domain in Four and a half LIM domains protein 2
Structural highlights
FunctionFHL2_HUMAN May function as a molecular transmitter linking various signaling pathways to transcriptional regulation. Negatively regulates the transcriptional repressor E4F1 and may function in cell growth. Inhibits the transcriptional activity of FOXO1 and its apoptotic function by enhancing the interaction of FOXO1 with SIRT1 and FOXO1 deacetylation.[1] [2] [3] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. See AlsoReferences
|
| ||||||||||||||||||