Lotem haleva/test page: Difference between revisions
Lotem Haleva (talk | contribs) No edit summary |
Michal Harel (talk | contribs) No edit summary |
||
| (11 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
==Trypsin== | ==Trypsin== | ||
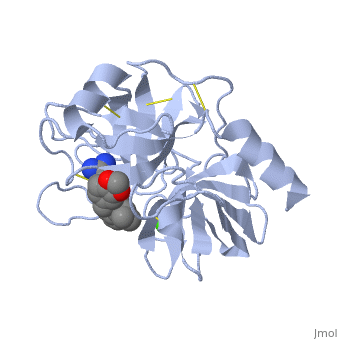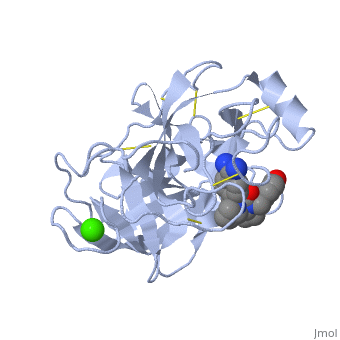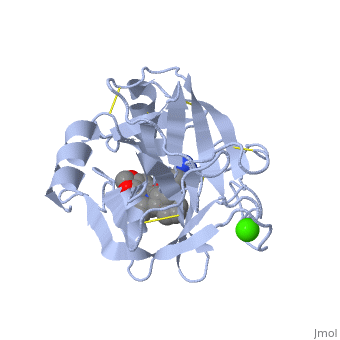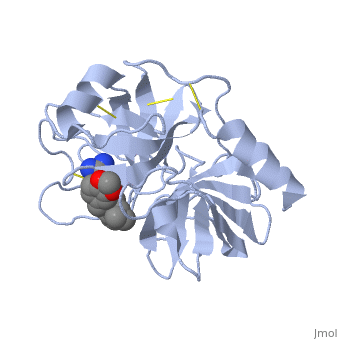
<StructureSection load='1y3v' size='340' side='right' caption=' | <StructureSection load='1y3v' size='340' side='right' caption='Bovine trypsine complex with inhibitor and Ca+2 ion (green) (PDB code [[1y3v]])' scene=''> | ||
Trypsin is a serine protease | '''Trypsin''' is a serine protease that found in the digestive system of many vertebrates, where it hydrolyses proteins<ref>doi:10.1016/0076-6879(94)44004-2</ref>. | ||
Trypsin is produced in the pancreas as the inactive protease trypsinogen. | Trypsin is produced in the pancreas as the inactive zymogen protease called trypsinogen. trypsinogen secreted into the small intestine where it activates trypsinogen into trypsin by the enzyme Antrookinaz using proteolytic decomposition. | ||
Trypsin Break peptide chains | Trypsin Break peptide chains at the carboxyl side of the amino acids lysine or arginine, | ||
except when | except when it is followed by proline. . | ||
== Function == | == Function == | ||
| Line 12: | Line 10: | ||
== Structure and Mechanism == | == Structure and Mechanism == | ||
Trypsin is a medium size globular protein that contains either <scene name='60/607865/Helix/1'>alpha helix</scene> | Trypsin is a medium size globular protein that contains either <scene name='60/607865/Helix/1'>alpha helix</scene> and <scene name='60/607865/Sheets/1'>beta sheets</scene>. | ||
The enzymatic mechanism of Trypsin is similar to the other serine proteases. These enzymes contain a catalytic triad consisting of <scene name='60/607865/Active_site/2'>histidine-57, aspartate-102, and serine-195</scene>, These three residues | The enzymatic mechanism of Trypsin is similar to the other serine proteases. These enzymes contain a catalytic triad consisting of <scene name='60/607865/Active_site/2'>histidine-57, aspartate-102, and serine-195</scene>, These three residues makes the active site serine nucleophilic <ref>doi:10.1007/s00018-005-5160-x</ref>. | ||
Trypsin contains an "oxyanion hole" formed by the backbone amide hydrogen atoms of <scene name='60/607865/Oxyanion_hole/1'>Gly-193 and Ser-195</scene> , which serves to stabilize the developing negative charge on the carbonyl oxygen atom of the cleaved amides. | Trypsin contains an "oxyanion hole" formed by the backbone amide hydrogen atoms of <scene name='60/607865/Oxyanion_hole/1'>Gly-193 and Ser-195</scene> , which serves to stabilize the developing negative charge on the carbonyl oxygen atom of the cleaved amides. | ||
The <scene name='60/607865/Asp_189/1'>aspartate residue</scene> located in the catalytic pocket of trypsin is responsible for attracting and stabilizing positively charged lysine or arginine, and as a result responsible for the specificity of the enzyme. | The <scene name='60/607865/Asp_189/1'>aspartate residue</scene> located in the catalytic pocket of trypsin is responsible for attracting and stabilizing positively charged lysine or arginine, and as a result responsible for the specificity of the enzyme <ref> doi:10.1021/pr0705035</ref>. | ||
== Structural highlights == | == Structural highlights == | ||
There are different ways to present the protein structure | There are different ways to present the protein structure :<scene name='60/607865/Balls_and_stik/1'>balls and stick</scene>, <scene name='60/607865/Surface/1'>surface</scene> and <scene name='60/607865/Backbone/1'>Backbone</scene>. Another way to view the protein is <scene name='60/607865/Rainbow_n_to_c/1'>Rainbow</scene> that show the protein from the N (blue) to C terminal (green). | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Latest revision as of 13:34, 9 January 2017
TrypsinTrypsin
Trypsin is a serine protease that found in the digestive system of many vertebrates, where it hydrolyses proteins[1]. Trypsin is produced in the pancreas as the inactive zymogen protease called trypsinogen. trypsinogen secreted into the small intestine where it activates trypsinogen into trypsin by the enzyme Antrookinaz using proteolytic decomposition. Trypsin Break peptide chains at the carboxyl side of the amino acids lysine or arginine, except when it is followed by proline. . FunctionTrypsin catalyzes the hydrolysis of peptide bonds and cutting proteins into smaller peptides in the duodenum. Structure and MechanismTrypsin is a medium size globular protein that contains either and . The enzymatic mechanism of Trypsin is similar to the other serine proteases. These enzymes contain a catalytic triad consisting of , These three residues makes the active site serine nucleophilic [2]. Trypsin contains an "oxyanion hole" formed by the backbone amide hydrogen atoms of , which serves to stabilize the developing negative charge on the carbonyl oxygen atom of the cleaved amides. The located in the catalytic pocket of trypsin is responsible for attracting and stabilizing positively charged lysine or arginine, and as a result responsible for the specificity of the enzyme [3].
Structural highlightsThere are different ways to present the protein structure :, and . Another way to view the protein is that show the protein from the N (blue) to C terminal (green).
|
| ||||||||||
ReferencesReferences
- ↑ Rawlings ND, Barrett AJ. Families of serine peptidases. Methods Enzymol. 1994;244:19-61. doi: 10.1016/0076-6879(94)44004-2. PMID:7845208 doi:http://dx.doi.org/10.1016/0076-6879(94)44004-2
- ↑ Polgar L. The catalytic triad of serine peptidases. Cell Mol Life Sci. 2005 Oct;62(19-20):2161-72. PMID:16003488 doi:http://dx.doi.org/10.1007/s00018-005-5160-x
- ↑ Rodriguez J, Gupta N, Smith RD, Pevzner PA. Does trypsin cut before proline? J Proteome Res. 2008 Jan;7(1):300-5. Epub 2007 Dec 8. PMID:18067249 doi:http://dx.doi.org/10.1021/pr0705035