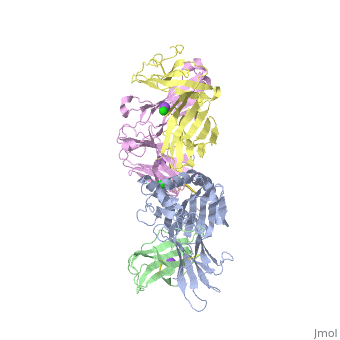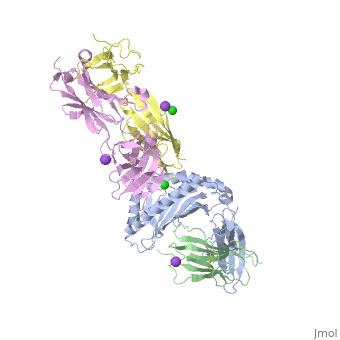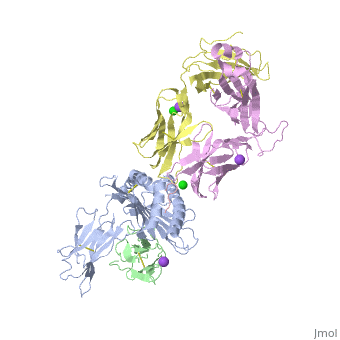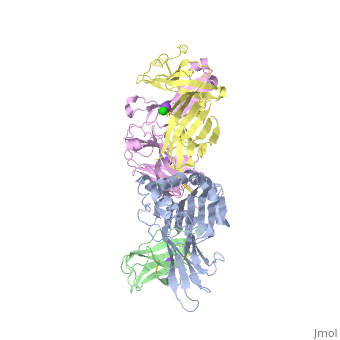4pri: Difference between revisions
m Protected "4pri" [edit=sysop:move=sysop] |
No edit summary |
||
| (8 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
The | ==Crystal structure of TK3 TCR-HLA-B*35:08-HPVG complex== | ||
<StructureSection load='4pri' size='340' side='right'caption='[[4pri]], [[Resolution|resolution]] 2.40Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[4pri]] is a 5 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens] and [https://en.wikipedia.org/wiki/Human_herpesvirus_4_strain_B95-8 Human herpesvirus 4 strain B95-8]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=4PRI OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=4PRI FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.4Å</td></tr> | |||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=CL:CHLORIDE+ION'>CL</scene>, <scene name='pdbligand=NA:SODIUM+ION'>NA</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=4pri FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4pri OCA], [https://pdbe.org/4pri PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=4pri RCSB], [https://www.ebi.ac.uk/pdbsum/4pri PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=4pri ProSAT]</span></td></tr> | |||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/C5MK56_HUMAN C5MK56_HUMAN] Involved in the presentation of foreign antigens to the immune system (By similarity).[SAAS:SAAS003006_004_004364] | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
Mutations within T cell epitopes represent a common mechanism of viral escape from the host protective immune response. The diverse T cell repertoire and the extensive human leukocyte antigen (HLA) polymorphism across populations is the evolutionary response to viral mutation. However, the molecular basis underpinning the interplay between HLA polymorphism, the T cell repertoire, and viral escape is unclear. Here we investigate the T cell response to a HLA-B*35:01- and HLA-B*35:08-restricted 407HPVGEADYFEY417 epitope from Epstein-Barr virus and naturally occurring variants at positions 4 and 5 thereof. Each viral variant differently impacted on the epitope's flexibility and conformation when bound to HLA-B*35:08 or HLA-B*35:01. We provide a molecular basis for understanding how the single residue polymorphism that discriminates between HLA-B*35:01/08 profoundly impacts on T cell receptor recognition. Surprisingly, one viral variant (P5-Glu to P5-Asp) effectively changed restriction preference from HLA-B*35:01 to HLA-B*35:08. Collectively, our study portrays the interplay between the T cell response, viral escape, and HLA polymorphism, whereby HLA polymorphism enables altered presentation of epitopes from different strains of Epstein-Barr virus. | |||
A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.,Liu YC, Chen Z, Neller MA, Miles JJ, Purcell AW, McCluskey J, Burrows SR, Rossjohn J, Gras S J Biol Chem. 2014 Jun 13;289(24):16688-16698. Epub 2014 Apr 23. PMID:24759101<ref>PMID:24759101</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 4pri" style="background-color:#fffaf0;"></div> | |||
==See Also== | |||
*[[Beta-2 microglobulin 3D structures|Beta-2 microglobulin 3D structures]] | |||
*[[MHC 3D structures|MHC 3D structures]] | |||
*[[MHC I 3D structures|MHC I 3D structures]] | |||
*[[T-cell receptor 3D structures|T-cell receptor 3D structures]] | |||
== References == | |||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
[[Category: Homo sapiens]] | |||
[[Category: Human herpesvirus 4 strain B95-8]] | |||
[[Category: Large Structures]] | |||
[[Category: Gras S]] | |||
[[Category: Rossjohn J]] | |||
[[Category: Yu Chih L]] | |||
Latest revision as of 20:21, 20 September 2023
Crystal structure of TK3 TCR-HLA-B*35:08-HPVG complexCrystal structure of TK3 TCR-HLA-B*35:08-HPVG complex
Structural highlights
FunctionC5MK56_HUMAN Involved in the presentation of foreign antigens to the immune system (By similarity).[SAAS:SAAS003006_004_004364] Publication Abstract from PubMedMutations within T cell epitopes represent a common mechanism of viral escape from the host protective immune response. The diverse T cell repertoire and the extensive human leukocyte antigen (HLA) polymorphism across populations is the evolutionary response to viral mutation. However, the molecular basis underpinning the interplay between HLA polymorphism, the T cell repertoire, and viral escape is unclear. Here we investigate the T cell response to a HLA-B*35:01- and HLA-B*35:08-restricted 407HPVGEADYFEY417 epitope from Epstein-Barr virus and naturally occurring variants at positions 4 and 5 thereof. Each viral variant differently impacted on the epitope's flexibility and conformation when bound to HLA-B*35:08 or HLA-B*35:01. We provide a molecular basis for understanding how the single residue polymorphism that discriminates between HLA-B*35:01/08 profoundly impacts on T cell receptor recognition. Surprisingly, one viral variant (P5-Glu to P5-Asp) effectively changed restriction preference from HLA-B*35:01 to HLA-B*35:08. Collectively, our study portrays the interplay between the T cell response, viral escape, and HLA polymorphism, whereby HLA polymorphism enables altered presentation of epitopes from different strains of Epstein-Barr virus. A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.,Liu YC, Chen Z, Neller MA, Miles JJ, Purcell AW, McCluskey J, Burrows SR, Rossjohn J, Gras S J Biol Chem. 2014 Jun 13;289(24):16688-16698. Epub 2014 Apr 23. PMID:24759101[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See Also
References
|
| ||||||||||||||||||