4ald: Difference between revisions
Jump to navigation
Jump to search
New page: left|200px<br /> <applet load="4ald" size="450" color="white" frame="true" align="right" spinBox="true" caption="4ald, resolution 2.8Å" /> '''HUMAN MUSCLE FRUCTOS... |
No edit summary |
||
| (27 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
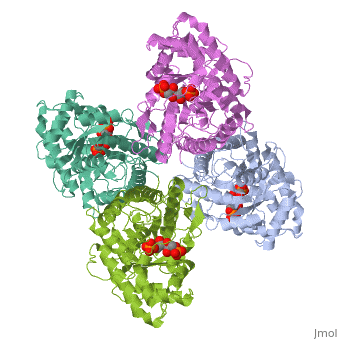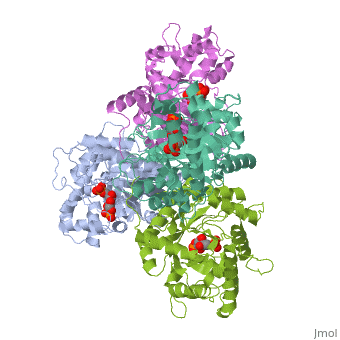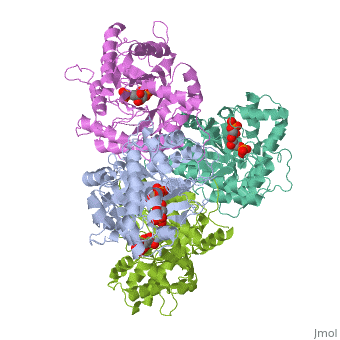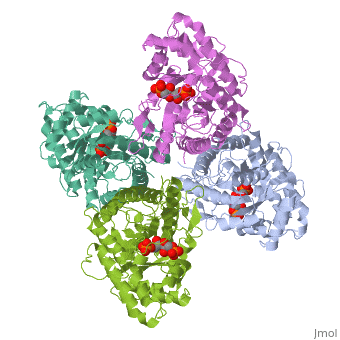
== | ==HUMAN MUSCLE FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE COMPLEXED WITH FRUCTOSE 1,6-BISPHOSPHATE== | ||
<StructureSection load='4ald' size='340' side='right'caption='[[4ald]], [[Resolution|resolution]] 2.80Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[4ald]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. The February 2004 RCSB PDB [https://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''The Glycolytic Enzymes'' by David S. Goodsell is [https://dx.doi.org/10.2210/rcsb_pdb/mom_2004_2 10.2210/rcsb_pdb/mom_2004_2]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=4ALD OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=4ALD FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.8Å</td></tr> | |||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=2FP:1,6-FRUCTOSE+DIPHOSPHATE+(LINEAR+FORM)'>2FP</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=4ald FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4ald OCA], [https://pdbe.org/4ald PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=4ald RCSB], [https://www.ebi.ac.uk/pdbsum/4ald PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=4ald ProSAT]</span></td></tr> | |||
</table> | |||
== Disease == | |||
[https://www.uniprot.org/uniprot/ALDOA_HUMAN ALDOA_HUMAN] Defects in ALDOA are the cause of glycogen storage disease type 12 (GSD12) [MIM:[https://omim.org/entry/611881 611881]; also known as red cell aldolase deficiency. A metabolic disorder associated with increased hepatic glycogen and hemolytic anemia. It may lead to myopathy with exercise intolerance and rhabdomyolysis.<ref>PMID:14766013</ref> <ref>PMID:2825199</ref> <ref>PMID:2229018</ref> <ref>PMID:8598869</ref> <ref>PMID:14615364</ref> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/ALDOA_HUMAN ALDOA_HUMAN] Plays a key role in glycolysis and gluconeogenesis. In addition, may also function as scaffolding protein (By similarity). | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/al/4ald_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=4ald ConSurf]. | |||
<div style="clear:both"></div> | |||
== | ==See Also== | ||
*[[Aldolase|Aldolase]] | |||
*[[Aldolase 3D structures|Aldolase 3D structures]] | |||
== | == References == | ||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
[[Category: | [[Category: Large Structures]] | ||
[[Category: RCSB PDB Molecule of the Month]] | |||
[[Category: The Glycolytic Enzymes]] | [[Category: The Glycolytic Enzymes]] | ||
[[Category: Dalby | [[Category: Dalby AR]] | ||
[[Category: Dauter | [[Category: Dauter Z]] | ||
[[Category: Littlechild | [[Category: Littlechild JA]] | ||
Latest revision as of 13:40, 1 March 2024
HUMAN MUSCLE FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE COMPLEXED WITH FRUCTOSE 1,6-BISPHOSPHATEHUMAN MUSCLE FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE COMPLEXED WITH FRUCTOSE 1,6-BISPHOSPHATE
Structural highlights
DiseaseALDOA_HUMAN Defects in ALDOA are the cause of glycogen storage disease type 12 (GSD12) [MIM:611881; also known as red cell aldolase deficiency. A metabolic disorder associated with increased hepatic glycogen and hemolytic anemia. It may lead to myopathy with exercise intolerance and rhabdomyolysis.[1] [2] [3] [4] [5] FunctionALDOA_HUMAN Plays a key role in glycolysis and gluconeogenesis. In addition, may also function as scaffolding protein (By similarity). Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. See AlsoReferences
|
| ||||||||||||||||||