Ribonuclease A Catalysis: Difference between revisions
No edit summary |
No edit summary |
||
| (26 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
< | <StructureSection load='7rsa' size='350' side='right' caption='' scene='Sandbox_Reserved_193/Rnasei_a/1 ' pspeed='8'> | ||
[[Image:RNaseAIII.png|300px|left|thumb|Figure I: Bovine Ribonuclease A. Colored residues are representative of amino acids important to both the acid base catalysis (Red: His12 and 119) and stabilization of the transition state (Blue: Lys41). Figure generated via ''Pymol'' ]] | |||
{{Clear}} | |||
= | |||
== | |||
[[Image: | |||
{{ | |||
===Acid Base Catalysis=== | ===Acid Base Catalysis=== | ||
In organic chemistry acid/base catalysis is the addition of an acid or base to accelerate a chemical reaction. Ribonuclease A, (RNase A), also uses acid/base catatalysis to chemically change its substrates. Acidic or basic residues of the enzyme transfer protons to or from the reactant in order to stabilize the developing charges in the transition state. The transfer of protons usually creates better leaving groups, making the reaction more energetically favorable. Histidine is a very common amino acid residue involved in cataylsis, as histidine has a pKa value close to neutral, (pKa=6); therefore, histidine can both accept and donate protons at physiological pH. | In organic chemistry acid/base catalysis is the addition of an acid or base to accelerate a chemical reaction. Ribonuclease A, (RNase A), also uses acid/base catatalysis to chemically change its substrates. Acidic or basic residues of the enzyme transfer protons to or from the reactant in order to stabilize the developing charges in the transition state. The transfer of protons usually creates better leaving groups, making the reaction more energetically favorable. Histidine is a very common amino acid residue involved in cataylsis, as histidine has a pKa value close to neutral, (pKa=6); therefore, histidine can both accept and donate protons at physiological pH. | ||
Acid/base catalysis by an enzyme is dependent on the pH of the environment and the pKa's of their residues. The pKa value will increase for an acidic residue if the environment is hydrophobic or if the adjacent residues are of similar charges. In the same environmental conditions, a basic residue will decrease the pKa. | Acid/base catalysis by an enzyme is dependent on the pH of the environment and the pKa's of their residues. The pKa value will increase for an acidic residue if the environment is hydrophobic or if the adjacent residues are of similar charges. In the same environmental conditions, a basic residue will decrease the pKa. This ability to alter the pKa of certain residues such as histidines, increases the diversity of reactions that an enzyme can perform. | ||
===Active Site Structure=== | ===Active Site Structure=== | ||
RNase A uses acid/base catlysis to speed up RNA hydrolysis. This occurs in the active site which is found in the cleft of RNase A and is the location of the chemical change in bound substrates. Subsites lining the active site cleft are important to the binding of single stranded RNA. Large quantities of positively charged residues, such as '' | RNase A uses acid/base catlysis to speed up RNA hydrolysis. This occurs in the <scene name='Sandbox_Reserved_193/Active_site_a/1'>active site</scene> which is found in the cleft of RNase A and is the location of the chemical change in bound substrates. Subsites lining the active site cleft are important to the binding of single stranded RNA. Large quantities of positively charged residues, such as <scene name='Sandbox_Reserved_193/Lys7_a/3'>Lys7</scene> and <scene name='Sandbox_Reserved_193/Lys66_a/2'>Lys66</scene> and <scene name='Sandbox_Reserved_193/Lys66_a/3'>Arg10</scene>, recognize the negative charge on the phosphate back bone of the RNA strand <ref name="Wlodrawer1988" />. | ||
The active site for RNase A, although fairly nonspecific, has some specificity for sites RNA hydrolysis. '' | The active site for RNase A, although fairly nonspecific, has some specificity for sites RNA hydrolysis. <scene name='Sandbox_Reserved_193/Thr45_a/1'>Threonine 45</scene>, located next to the active site, will hydrogen bond to pyrimidine bases, but sterically hinder the binding of a purine on the 5' strand of OH. Thr45 significantly decreases the rate of hydrolysis of polymeric purine strands, such as poly A, by a thousand fold, as compared to polymeric pyrimidine strands <ref name="Wlodrawer1988" />. | ||
Early studies on RNase A catalysis showed that alkylation of His12 and His119 significantly decreased its catalytic activity, prompting the hypothesis that these two histidines were the acid/base catalyst. Confirmation of this hypothesis came when these histidines were replaced with alanine and the reaction rates of either mutation dropped by ten-thousand fold. | Early studies on RNase A catalysis showed that alkylation of His12 and His119 significantly decreased its catalytic activity, prompting the hypothesis that these two histidines were the acid/base catalyst. Confirmation of this hypothesis came when these histidines were replaced with alanine and the reaction rates of either mutation dropped by ten-thousand fold <ref name="Wlodrawer1988" />. | ||
=='''Acid Base Catalysis by RNase A'''== | =='''Acid Base Catalysis by RNase A'''== | ||
===Mechanism=== | ===Mechanism=== | ||
RNase A catalyzes the cleavage of the Phosphodiester bonds in two steps: the formation of the pentavalent phosphate transition state and subsequent degradation of the 2’3’ cyclic phosphate intermediate. An important part of the reaction is the ability of histidine (His 12 and His119) to both accept and donate electrons, allowing these histidine to be an acid or a base, making the reaction pH dependent <ref name="Raines1998" />. | |||
| Line 30: | Line 24: | ||
RNA hydrolysis begins when <scene name='Sandbox_Reserved_193/ | RNA hydrolysis begins when <scene name='Sandbox_Reserved_193/His12a_a/1'>His12</scene> abstracts a proton from the 2’ OH group on RNA; thus, assisting in the nucleophilic attack of the 2’ oxygen on the electrophilic phosphorus atom. A transition state is then formed, having a pentavalent phosphate, which is stabilized by the positively charged amino group of <scene name='Sandbox_Reserved_193/Lys41a_a/1'>Lys41</scene> and the main chain amide nitrogen of Phe120. <scene name='Sandbox_Reserved_193/His119a_a/1'>His119</scene> then protonates the 5' oxygen on the ribose ring and the transition state falls to form a 2’3’cyclic phosphate intermediate <ref name="Raines1998" />. | ||
In a secondary and separate reaction, the 2’,3’ cyclic phosphate is hydrolyzed to a mixture of 2'phosphate and 3' hydroxyl. His12 donates a proton to the leaving group of this reaction, the 3’ oxygen of the cyclic intermediate. Simultaneously, His-119 abstracts the proton from a water molecule, activating it for nucleophilic attack. The activated water molecule attacks the cyclic phosphate causing the cleavage of the 2'3’ cyclic phosphate intermediate. The truncated nucleotide is then released with a 3’ phosphate group. | In a secondary and separate reaction, the 2’,3’ cyclic phosphate is hydrolyzed to a mixture of 2'phosphate and 3' hydroxyl. His12 donates a proton to the leaving group of this reaction, the 3’ oxygen of the cyclic intermediate. Simultaneously, His-119 abstracts the proton from a water molecule, activating it for nucleophilic attack. The activated water molecule attacks the cyclic phosphate causing the cleavage of the 2'3’ cyclic phosphate intermediate. The truncated nucleotide is then released with a 3’ phosphate group <ref name="Raines1998" />. | ||
===Inhibitors=== | ===Inhibitors=== | ||
Due to the highly catalytic nature of RNase A for RNA strands, mammalian cells have developed a protective inhibitor to prevent pancreatic ribonucleases from degrading cystolic RNA. Ribonuclease Inhibitor (RI) tightly associates to the active site due to its non-globular nature. ''' | Due to the highly catalytic nature of RNase A for RNA strands, mammalian cells have developed a protective inhibitor to prevent pancreatic ribonucleases from degrading cystolic RNA. Ribonuclease Inhibitor (RI) tightly associates to the active site due to its non-globular nature. RI is a large 50 kD protein that is composed of 16 repeating alpha and beta chains, giving it a noticable horseshoe like appearance. It has been suggested that RI has the highest protein-protein interactions with an approximate dissociation constant (Kd) of 5.8 X 10-14 for almost all types of RNases <ref name="Vicentini1996" />. | ||
[[Image:RI.PNG|300px|Right|thumb|Figure III: Ribonuclease Inhibitor-RNase A Complex. Left, Ribonuclease Inhibitor (RI)is composed of alternating alpha helix (blue) and beta sheets (green). Right, RI-RNase A inhibition forms when RI complex with the active site cleft of RNase (yellow). Figure generated via ''Pymol'']] | |||
The ability to be selective for almost all types of RNases, and yet retain such a high Kd is product of its mechanism. The interior residues of the horseshoe shaped RI are able to bind to the charged residues of the active site cleft of RNase A, such as Lys7, Lys9, Lys 41 and Gln11. By studying the amphibian RNase, Onconase, the residues Lys7 and Gln11 of RNase A were shown to be the most important in this interaction. In onconase, these residues are replaced with non-charged amino acids, which help prevent the binding of RI to the protein <ref name="Turcotte2008" />. | |||
</StructureSection> | |||
=References= | |||
{{Reflist| refs= | |||
<ref name="Raines1998">Raines, R. Ribonuclease A. ''Chemistry Review'': (1998) Vol. 98 pp. 1045-1068</ref> | |||
<ref name="Turcotte2008">Turcotte, R., Raines, R., Interactions of Onconase with Human Ribonuclease Inhibitor. ''Biochemical Biophysical Research Communities'':(2008) Vol. 377, Iss. 4, pp. 512-414</ref> | |||
<ref name="Vicentini1996"> Vicentini,A., Protein Chemical and Kinetic Characterization of Recombinant Porcine Ribonuclease Inhibitor Expressed in ''Saccharomyces cerevisae''. ''Biochemistry'': (1996) Vol. 29, pp.8827-8834</ref> | |||
<ref name="Wlodrawer1988">Wlodrawer, A., Svensson, L., Sjohin, L., Gilliland, G. Structure of Phosphate-Free Ribonuclease A Refined at 1.26A. ''Biochemistry'':(1988) Vol. 27 pp. 2705-2717</ref> | |||
}} | |||
=Related Sites= | =Related Sites= | ||
http://en.wikipedia.org/wiki/RNase_A | |||
http://en.wikipedia.org/wiki/Ribonuclease_inhibitor | |||
Latest revision as of 12:46, 8 December 2019
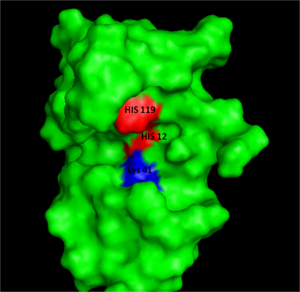 Acid Base CatalysisIn organic chemistry acid/base catalysis is the addition of an acid or base to accelerate a chemical reaction. Ribonuclease A, (RNase A), also uses acid/base catatalysis to chemically change its substrates. Acidic or basic residues of the enzyme transfer protons to or from the reactant in order to stabilize the developing charges in the transition state. The transfer of protons usually creates better leaving groups, making the reaction more energetically favorable. Histidine is a very common amino acid residue involved in cataylsis, as histidine has a pKa value close to neutral, (pKa=6); therefore, histidine can both accept and donate protons at physiological pH. Acid/base catalysis by an enzyme is dependent on the pH of the environment and the pKa's of their residues. The pKa value will increase for an acidic residue if the environment is hydrophobic or if the adjacent residues are of similar charges. In the same environmental conditions, a basic residue will decrease the pKa. This ability to alter the pKa of certain residues such as histidines, increases the diversity of reactions that an enzyme can perform.
Active Site StructureRNase A uses acid/base catlysis to speed up RNA hydrolysis. This occurs in the which is found in the cleft of RNase A and is the location of the chemical change in bound substrates. Subsites lining the active site cleft are important to the binding of single stranded RNA. Large quantities of positively charged residues, such as and and , recognize the negative charge on the phosphate back bone of the RNA strand [1]. The active site for RNase A, although fairly nonspecific, has some specificity for sites RNA hydrolysis. , located next to the active site, will hydrogen bond to pyrimidine bases, but sterically hinder the binding of a purine on the 5' strand of OH. Thr45 significantly decreases the rate of hydrolysis of polymeric purine strands, such as poly A, by a thousand fold, as compared to polymeric pyrimidine strands [1]. Early studies on RNase A catalysis showed that alkylation of His12 and His119 significantly decreased its catalytic activity, prompting the hypothesis that these two histidines were the acid/base catalyst. Confirmation of this hypothesis came when these histidines were replaced with alanine and the reaction rates of either mutation dropped by ten-thousand fold [1].
Acid Base Catalysis by RNase AMechanismRNase A catalyzes the cleavage of the Phosphodiester bonds in two steps: the formation of the pentavalent phosphate transition state and subsequent degradation of the 2’3’ cyclic phosphate intermediate. An important part of the reaction is the ability of histidine (His 12 and His119) to both accept and donate electrons, allowing these histidine to be an acid or a base, making the reaction pH dependent [2].

RNA hydrolysis begins when abstracts a proton from the 2’ OH group on RNA; thus, assisting in the nucleophilic attack of the 2’ oxygen on the electrophilic phosphorus atom. A transition state is then formed, having a pentavalent phosphate, which is stabilized by the positively charged amino group of and the main chain amide nitrogen of Phe120. then protonates the 5' oxygen on the ribose ring and the transition state falls to form a 2’3’cyclic phosphate intermediate [2].
In a secondary and separate reaction, the 2’,3’ cyclic phosphate is hydrolyzed to a mixture of 2'phosphate and 3' hydroxyl. His12 donates a proton to the leaving group of this reaction, the 3’ oxygen of the cyclic intermediate. Simultaneously, His-119 abstracts the proton from a water molecule, activating it for nucleophilic attack. The activated water molecule attacks the cyclic phosphate causing the cleavage of the 2'3’ cyclic phosphate intermediate. The truncated nucleotide is then released with a 3’ phosphate group [2].
InhibitorsDue to the highly catalytic nature of RNase A for RNA strands, mammalian cells have developed a protective inhibitor to prevent pancreatic ribonucleases from degrading cystolic RNA. Ribonuclease Inhibitor (RI) tightly associates to the active site due to its non-globular nature. RI is a large 50 kD protein that is composed of 16 repeating alpha and beta chains, giving it a noticable horseshoe like appearance. It has been suggested that RI has the highest protein-protein interactions with an approximate dissociation constant (Kd) of 5.8 X 10-14 for almost all types of RNases [3].  The ability to be selective for almost all types of RNases, and yet retain such a high Kd is product of its mechanism. The interior residues of the horseshoe shaped RI are able to bind to the charged residues of the active site cleft of RNase A, such as Lys7, Lys9, Lys 41 and Gln11. By studying the amphibian RNase, Onconase, the residues Lys7 and Gln11 of RNase A were shown to be the most important in this interaction. In onconase, these residues are replaced with non-charged amino acids, which help prevent the binding of RI to the protein [4].
|
| ||||||||||
ReferencesReferences
- ↑ 1.0 1.1 1.2 Cite error: Invalid
<ref>tag; no text was provided for refs namedWlodrawer1988 - ↑ 2.0 2.1 2.2 Cite error: Invalid
<ref>tag; no text was provided for refs namedRaines1998 - ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedVicentini1996 - ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedTurcotte2008