1rcx: Difference between revisions
No edit summary |
No edit summary |
||
| (23 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
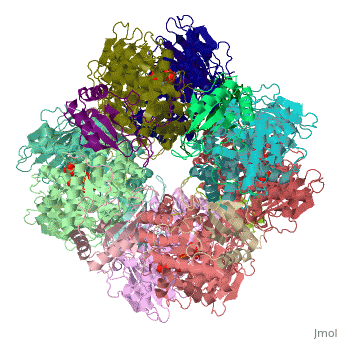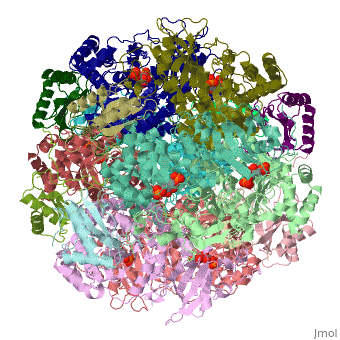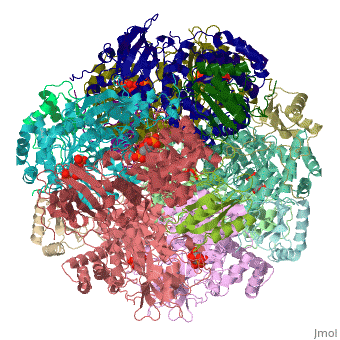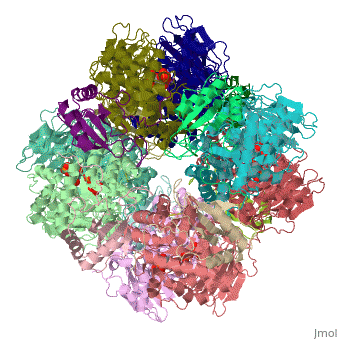
== | ==NON-ACTIVATED SPINACH RUBISCO IN COMPLEX WITH ITS SUBSTRATE RIBULOSE-1,5-BISPHOSPHATE== | ||
The three-dimensional structure of the complex of ribulose | <StructureSection load='1rcx' size='340' side='right'caption='[[1rcx]], [[Resolution|resolution]] 2.40Å' scene=''> | ||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[1rcx]] is a 16 chain structure with sequence from [https://en.wikipedia.org/wiki/Spinacia_oleracea Spinacia oleracea]. The November 2000 RCSB PDB [https://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''Rubisco'' by David S. Goodsell is [https://dx.doi.org/10.2210/rcsb_pdb/mom_2000_11 10.2210/rcsb_pdb/mom_2000_11]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1RCX OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1RCX FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.4Å</td></tr> | |||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=RUB:RIBULOSE-1,5-DIPHOSPHATE'>RUB</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1rcx FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1rcx OCA], [https://pdbe.org/1rcx PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1rcx RCSB], [https://www.ebi.ac.uk/pdbsum/1rcx PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1rcx ProSAT]</span></td></tr> | |||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/RBL_SPIOL RBL_SPIOL] RuBisCO catalyzes two reactions: the carboxylation of D-ribulose 1,5-bisphosphate, the primary event in carbon dioxide fixation, as well as the oxidative fragmentation of the pentose substrate in the photorespiration process. Both reactions occur simultaneously and in competition at the same active site.[HAMAP-Rule:MF_01338] | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/rc/1rcx_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1rcx ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
The three-dimensional structure of the complex of ribulose 1,5-bisphosphate carboxylase/oxygenase (rubisco; EC 4.1.1.39) from spinach with its natural substrate ribulose 1,5-bisphosphate (RuBP) has been determined both under activating and non-activating conditions by X-ray crystallography to a resolution of 2.1 A and 2.4 A, respectively. Under activating conditions, the use of calcium instead of magnesium as the activator metal ion enabled us to trap the substrate in a stable complex for crystallographic analysis. Comparison of the structure of the activated and the non-activated RuBP complexes shows a tighter binding for the substrate in the non-activated form of the enzyme, in line with previous solution studies. In the non-activated complex, the substrate triggers isolation of the active site by inducing movements of flexible loop regions of the catalytic subunits. In contrast, in the activated complex the active site remains partly open, probably awaiting the binding of the gaseous substrate. By inspection of the structures and by comparison with other complexes of the enzyme we were able to identify a network of hydrogen bonds that stabilise a closed active site structure during crucial steps in the reaction. The present structure underlines the central role of the carbamylated lysine 201 in both activation and catalysis, and completes available structural information for our proposal on the mechanism of the enzyme. | |||
The structure of the complex between rubisco and its natural substrate ribulose 1,5-bisphosphate.,Taylor TC, Andersson I J Mol Biol. 1997 Jan 31;265(4):432-44. PMID:9034362<ref>PMID:9034362</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
[[Category: | <div class="pdbe-citations 1rcx" style="background-color:#fffaf0;"></div> | ||
[[Category: | |||
==See Also== | |||
*[[RuBisCO|RuBisCO]] | |||
*[[RuBisCO 3D structures|RuBisCO 3D structures]] | |||
== References == | |||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
[[Category: Large Structures]] | |||
[[Category: RCSB PDB Molecule of the Month]] | |||
[[Category: Rubisco]] | [[Category: Rubisco]] | ||
[[Category: Spinacia oleracea]] | [[Category: Spinacia oleracea]] | ||
[[Category: Andersson | [[Category: Andersson I]] | ||
[[Category: Taylor | [[Category: Taylor TC]] | ||
Latest revision as of 10:18, 30 October 2024
NON-ACTIVATED SPINACH RUBISCO IN COMPLEX WITH ITS SUBSTRATE RIBULOSE-1,5-BISPHOSPHATENON-ACTIVATED SPINACH RUBISCO IN COMPLEX WITH ITS SUBSTRATE RIBULOSE-1,5-BISPHOSPHATE
Structural highlights
FunctionRBL_SPIOL RuBisCO catalyzes two reactions: the carboxylation of D-ribulose 1,5-bisphosphate, the primary event in carbon dioxide fixation, as well as the oxidative fragmentation of the pentose substrate in the photorespiration process. Both reactions occur simultaneously and in competition at the same active site.[HAMAP-Rule:MF_01338] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedThe three-dimensional structure of the complex of ribulose 1,5-bisphosphate carboxylase/oxygenase (rubisco; EC 4.1.1.39) from spinach with its natural substrate ribulose 1,5-bisphosphate (RuBP) has been determined both under activating and non-activating conditions by X-ray crystallography to a resolution of 2.1 A and 2.4 A, respectively. Under activating conditions, the use of calcium instead of magnesium as the activator metal ion enabled us to trap the substrate in a stable complex for crystallographic analysis. Comparison of the structure of the activated and the non-activated RuBP complexes shows a tighter binding for the substrate in the non-activated form of the enzyme, in line with previous solution studies. In the non-activated complex, the substrate triggers isolation of the active site by inducing movements of flexible loop regions of the catalytic subunits. In contrast, in the activated complex the active site remains partly open, probably awaiting the binding of the gaseous substrate. By inspection of the structures and by comparison with other complexes of the enzyme we were able to identify a network of hydrogen bonds that stabilise a closed active site structure during crucial steps in the reaction. The present structure underlines the central role of the carbamylated lysine 201 in both activation and catalysis, and completes available structural information for our proposal on the mechanism of the enzyme. The structure of the complex between rubisco and its natural substrate ribulose 1,5-bisphosphate.,Taylor TC, Andersson I J Mol Biol. 1997 Jan 31;265(4):432-44. PMID:9034362[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||