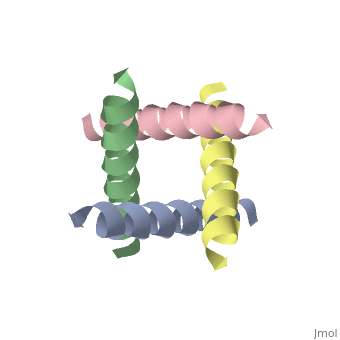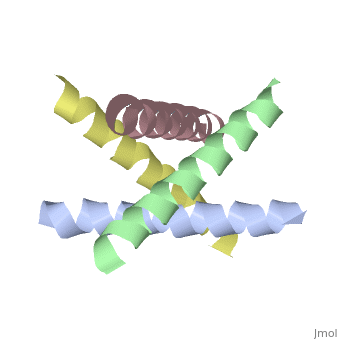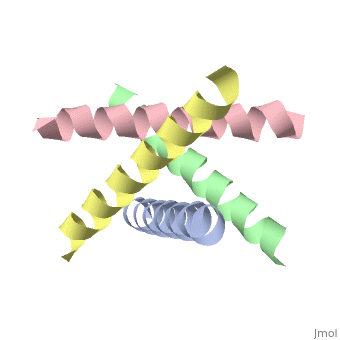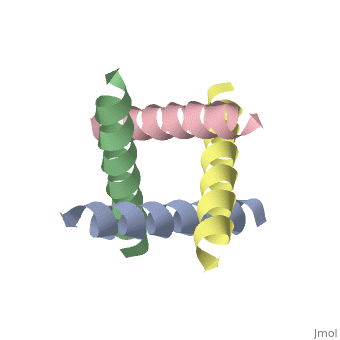M2 Proton Channel: Difference between revisions
Sarah Henke (talk | contribs) |
No edit summary |
||
| (36 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
<StructureSection load='1nyj' size='340' side='right' caption='The closed state structure of M2 protein H+ channel by solid state NMR spectroscopy ([[1nyj]])' scene=''> | |||
== M2 Proton Channel from ''Influenza'' A Virus == | == M2 Proton Channel from ''Influenza'' A Virus == | ||
< | == Background == | ||
The M2 proton channel is a key protein that leads to viral infection.<ref name="Takeuchi" /> The M2 proton channel acidifies the virion which allows the viral matrix protein (M1) to disassociate from the ribonucleoprotein (RNP).<ref name="Wu">PMID:12972147 </ref> This allows the RNP to be transported to the nucleus of the cell. Several recent studies have looked at the effects of <scene name='User:Sarah_Henke/Sandbox_1/Amantadine/1'>amantadine</scene> ([[Symmetrel]])<ref name="Stouffer">PMID:18235504 </ref> and <scene name='User:Sarah_Henke/Sandbox_1/Rimantadine/1'>rimantadine</scene> ([[Flumadine]])<ref name="Schnell">PMID:18235503 </ref> on inhibiting the transfer of protons through the M2 channel.<ref name="Stouffer" /> Amantadine is a proton surrogate that competes with protons for binding to His37, the residue involved in the gating mechanism.<ref name="Lear" /><ref>PMID:3662473</ref><ref>PMID:17156962</ref> It has been found that M2 is resistant to these two drugs in 90% of humans, birds and pigs. Understanding the structure and function of this proton channel is necessary in solving the resistance problem.<ref name="Stouffer" /> | |||
== | == Animation of Opening and Closing == | ||
A [[morph]] animation of the H2 proton channel opening and closing is available at [[Proton Channels]]. | |||
== Structure == | == Structure == | ||
The M2 proton channel from influenza A is 97 amino acid residues and forms a 24-residue N-terminal extracellular domain, a 19-residue trans-membrane domain, and a 54-residue C-terminal cytoplasmic domain.<ref name="Wu" /> The 19-residue TM domain forms the highly selective proton channel.<ref name="Takeuchi">PMID:12972149 </ref> Circular dichroism spectra has shown the TM domain to form an <scene name='User:Sarah_Henke/Sandbox_1/Momomer/2'>α-helix</scene> that spans the membrane.<ref name="Wu" /> By analytical ultracentrifugation, the TM domain is found to form <scene name='User:Sarah_Henke/Sandbox_1/Alpha_hlix/1'>homotetramers</scene> which contains four identical α-helices.<ref name="Takeuchi" /> Secondary structure is coded by the following, if present: {{Template:ColorKey_Helix}}, {{Template:ColorKey_Strand}}, and {{Template:ColorKey_Turn}}. When viewed in the <scene name='User:Sarah_Henke/Sandbox_1/N_to_c/1'>N->C color coding</scene> the <FONT COLOR="blue">'''N-terminus'''</FONT> is located near the external side of the membrane while the <FONT COLOR="red">'''C-terminus'''</FONT> is located near the internal side of the membrane, closest to the virion. This tetrameric bundle of the TM domain is found by NMR to be tilted by 25-38° from the channel axis. | The M2 proton channel from influenza A is 97 amino acid residues and forms a 24-residue N-terminal extracellular domain, a 19-residue trans-membrane domain, and a 54-residue C-terminal cytoplasmic domain.<ref name="Wu" /> The 19-residue TM domain forms the highly selective proton channel.<ref name="Takeuchi">PMID:12972149 </ref> Circular dichroism spectra has shown the TM domain to form an <scene name='User:Sarah_Henke/Sandbox_1/Momomer/2'>α-helix</scene> that spans the membrane.<ref name="Wu" /> By analytical ultracentrifugation, the TM domain is found to form <scene name='User:Sarah_Henke/Sandbox_1/Alpha_hlix/1'>homotetramers</scene> which contains four identical α-helices.<ref name="Takeuchi" /> Secondary structure is color coded by the following, if present: {{Template:ColorKey_Helix}}, {{Template:ColorKey_Strand}}, and {{Template:ColorKey_Turn}}. When viewed in the <scene name='User:Sarah_Henke/Sandbox_1/N_to_c/1'>N->C color coding</scene> the <FONT COLOR="blue">'''N-terminus'''</FONT> (see scale below) is located near the external side of the membrane while the <FONT COLOR="red">'''C-terminus'''</FONT> is located near the internal side of the membrane, closest to the virion. This tetrameric bundle of the TM domain is found by NMR to be tilted by 25-38° from the channel axis. The tetrameric helices form a left-handed bundle that resembles a truncated cone. The TM helicies are arranged around the channel pore with an approximate four-fold rotational symmetry.<ref name="Takeuchi" /> | ||
{{Template:ColorKey_N2CRainbow}} | {{Template:ColorKey_N2CRainbow}} | ||
== Central Cavity == | == Central Cavity == | ||
The hydrophilic residues in each α-helix monomer are oriented towards the pore lumen. | The hydrophilic residues in each α-helix monomer are oriented towards the pore lumen. The <scene name='User:Sarah_Henke/Sandbox_1/Hydrophobic/2'>hydrophobic</scene> residues will be in contact with the membrane (Color code= {{Template:ColorKey_Hydrophobic}} or {{Template:ColorKey_Polar}}). Most of the residues in the M2 channel are hydrophobic except Ser31 Gly34, and His37.<ref name="Wu" /> The central cavity of the M2 proton channel is most constricted near residue <scene name='User:Sarah_Henke/Sandbox_1/Val27/1'>Val27</scene>. After this residue, the cavity opens to a water-filled pore that is lined with residues <scene name='User:Sarah_Henke/Sandbox_1/Pore/1'>Ala30, Ser31, and Gly34</scene>.<ref name="Stouffer" /> Mutagenesis studies have found that the residues facing the pore are Val27, Ala30, Ser31, Gly34, His37, Leu38, and Trp41.<ref name="Wu" /> The central cavity also constricts at residues His37 and Trp41.<ref name="Stouffer" /> Residues <scene name='User:Sarah_Henke/Sandbox_1/His_37/1'>His37</scene> and <scene name='User:Sarah_Henke/Sandbox_1/Trp20/1'>Trp41</scene> play a key role in the gating mechanism.<ref name="Wu" /> In the closed state, the <scene name='User:Sarah_Henke/Sandbox_1/Histrp/1'>His37 and Trp41</scene> residues block the channel, preventing proton conductance. <ref name="Lear">PMID:12972146 </ref> | ||
== pH Gating == | == pH Gating == | ||
The M2 channel is low-pH gated and has a 50-fold increase in proton conductance when the pH drops from 8.2 down to 4.2.<ref name="Wu" /> The His side chain, a five-membered ring, has two nitrogen atoms. | The M2 channel is low-pH gated and has a 50-fold increase in proton conductance when the pH drops from 8.2 down to 4.2.<ref name="Wu" /> The His side chain, a five-membered ring, has two nitrogen atoms. At a pH of 7 or higher, only one nitrogen is protonated in the neutral imidazole form. However, at a pH of about 5.8, the second nitrogen in one or two helicies becomes protonated and forms the cationic imidazolium form. This protonation causes the His37 residues to move away from each other due to electrostatic repulsion.<ref name="Takeuchi" /><ref name="Lear" /><ref name="Wu" /> The bi-protonated His37 residues are then stabilized by the Trp41 residues by a cation-π interaction. This allows the channel to open and allow water from the pore to penetrate.<ref name="Lear" /> This forms what is called a proton-conductive water wire through the gate.<ref name="Wu" /> | ||
== Selectivity == | == Selectivity == | ||
Because the N-terminus is very constricted near Val27, the M2 channel is highly selective for protons. | Because the N-terminus is very constricted near Val27, the M2 channel is highly selective for protons. The restricted N-terminus only allows protons to penetrate into the aqueous pore through hydrogen-bonded chains of water. Therefore, it would be extremely difficult for hydrated sodium or potassium to penetrate the restricted areas of the M2 channel.<ref name="Stouffer" /> | ||
==3D structures of proton channel== | |||
See proton channel in [[Ion channels]] | |||
==Additional Resources== | |||
*[[Proton Channels]] includes a morph animation of the H2 proton channel opening and closing. | |||
*[[Membrane Channels & Pumps]] | |||
<br /> | |||
*[[Rimantadine]]<br /> | |||
*[[Amantadine]]<br /> | |||
*[[Treatments:M2 Proton Channel Inhibitor Pharmacokinetics]]<br /> | |||
*[[Treatments:Influenza]]. | |||
</StructureSection> | |||
== References == | == References == | ||
<references /> | <references /> | ||
Latest revision as of 12:12, 14 January 2024
M2 Proton Channel from Influenza A VirusBackgroundThe M2 proton channel is a key protein that leads to viral infection.[1] The M2 proton channel acidifies the virion which allows the viral matrix protein (M1) to disassociate from the ribonucleoprotein (RNP).[2] This allows the RNP to be transported to the nucleus of the cell. Several recent studies have looked at the effects of (Symmetrel)[3] and (Flumadine)[4] on inhibiting the transfer of protons through the M2 channel.[3] Amantadine is a proton surrogate that competes with protons for binding to His37, the residue involved in the gating mechanism.[5][6][7] It has been found that M2 is resistant to these two drugs in 90% of humans, birds and pigs. Understanding the structure and function of this proton channel is necessary in solving the resistance problem.[3] Animation of Opening and ClosingA morph animation of the H2 proton channel opening and closing is available at Proton Channels. StructureThe M2 proton channel from influenza A is 97 amino acid residues and forms a 24-residue N-terminal extracellular domain, a 19-residue trans-membrane domain, and a 54-residue C-terminal cytoplasmic domain.[2] The 19-residue TM domain forms the highly selective proton channel.[1] Circular dichroism spectra has shown the TM domain to form an that spans the membrane.[2] By analytical ultracentrifugation, the TM domain is found to form which contains four identical α-helices.[1] Secondary structure is color coded by the following, if present: Alpha Helices, Beta Strands , and Turns. When viewed in the the N-terminus (see scale below) is located near the external side of the membrane while the C-terminus is located near the internal side of the membrane, closest to the virion. This tetrameric bundle of the TM domain is found by NMR to be tilted by 25-38° from the channel axis. The tetrameric helices form a left-handed bundle that resembles a truncated cone. The TM helicies are arranged around the channel pore with an approximate four-fold rotational symmetry.[1]
Central CavityThe hydrophilic residues in each α-helix monomer are oriented towards the pore lumen. The residues will be in contact with the membrane (Color code= Hydrophobic or Polar). Most of the residues in the M2 channel are hydrophobic except Ser31 Gly34, and His37.[2] The central cavity of the M2 proton channel is most constricted near residue . After this residue, the cavity opens to a water-filled pore that is lined with residues .[3] Mutagenesis studies have found that the residues facing the pore are Val27, Ala30, Ser31, Gly34, His37, Leu38, and Trp41.[2] The central cavity also constricts at residues His37 and Trp41.[3] Residues and play a key role in the gating mechanism.[2] In the closed state, the residues block the channel, preventing proton conductance. [5] pH GatingThe M2 channel is low-pH gated and has a 50-fold increase in proton conductance when the pH drops from 8.2 down to 4.2.[2] The His side chain, a five-membered ring, has two nitrogen atoms. At a pH of 7 or higher, only one nitrogen is protonated in the neutral imidazole form. However, at a pH of about 5.8, the second nitrogen in one or two helicies becomes protonated and forms the cationic imidazolium form. This protonation causes the His37 residues to move away from each other due to electrostatic repulsion.[1][5][2] The bi-protonated His37 residues are then stabilized by the Trp41 residues by a cation-π interaction. This allows the channel to open and allow water from the pore to penetrate.[5] This forms what is called a proton-conductive water wire through the gate.[2] SelectivityBecause the N-terminus is very constricted near Val27, the M2 channel is highly selective for protons. The restricted N-terminus only allows protons to penetrate into the aqueous pore through hydrogen-bonded chains of water. Therefore, it would be extremely difficult for hydrated sodium or potassium to penetrate the restricted areas of the M2 channel.[3] 3D structures of proton channelSee proton channel in Ion channels Additional Resources
|
| |||||||||||||||||||
ReferencesReferences
- ↑ 1.0 1.1 1.2 1.3 1.4 Takeuchi H, Okada A, Miura T. Roles of the histidine and tryptophan side chains in the M2 proton channel from influenza A virus. FEBS Lett. 2003 Sep 18;552(1):35-8. PMID:12972149
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 Wu Y, Voth GA. Computational studies of proton transport through the M2 channel. FEBS Lett. 2003 Sep 18;552(1):23-7. PMID:12972147
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, Soto CS, Tereshko V, Nanda V, Stayrook S, DeGrado WF. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008 Jan 31;451(7178):596-9. PMID:18235504 doi:10.1038/nature06528
- ↑ Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008 Jan 31;451(7178):591-5. PMID:18235503 doi:10.1038/nature06531
- ↑ 5.0 5.1 5.2 5.3 Lear JD. Proton conduction through the M2 protein of the influenza A virus; a quantitative, mechanistic analysis of experimental data. FEBS Lett. 2003 Sep 18;552(1):17-22. PMID:12972146
- ↑ Anderson EL, Van Voris LP, Bartram J, Hoffman HE, Belshe RB. Pharmacokinetics of a single dose of rimantadine in young adults and children. Antimicrob Agents Chemother. 1987 Jul;31(7):1140-2. PMID:3662473
- ↑ Wang P, Liang YZ, Chen BM, Zhou N, Yi LZ, Yu Y, Yi ZB. Quantitative determination of amantadine in human plasma by liquid chromatography-mass spectrometry and the application in a bioequivalence study. J Pharm Biomed Anal. 2007 Mar 12;43(4):1519-25. Epub 2006 Dec 6. PMID:17156962 doi:10.1016/j.jpba.2006.10.044