Chemotaxis protein: Difference between revisions
Jump to navigation
Jump to search
Michal Harel (talk | contribs) No edit summary |
Michal Harel (talk | contribs) No edit summary |
||
| (37 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
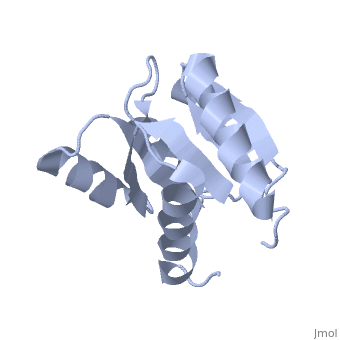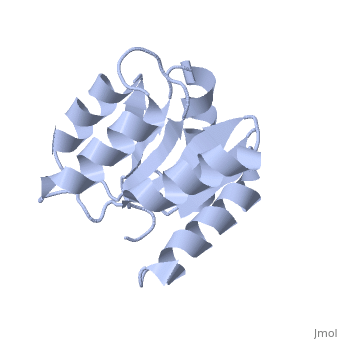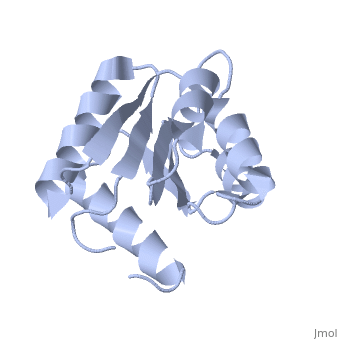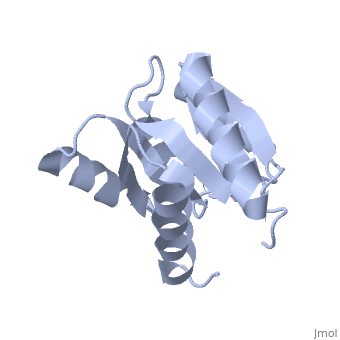
<StructureSection load='1e6m' size='350' side='right' caption='E. coli CheY (PDB entry [[1e6m]])' scene=''> | |||
'''Chemotaxis proteins''' direct cell movement according to certain chemicals in their environment.<ref>PMID:4598187</ref> | |||
*'''CheY''' is a response regulator determining the swimming behaviour of the cell<ref>PMID:7623663</ref>. | |||
*'''CheR''' is a SAM-dependent methyltransferase which binds to the C-terminus of some chemotaxis receptors and methylates neighbouring receptor molecules<ref>PMID:9628482</ref>. See [[Molecular Playground/CheR]] and [[Chemotaxis receptor methyltransferase]]. | |||
*'''CheA''' is a histidine kinase which donates phosphoric groups to CheB and CheY to control swimming behaviour<ref>PMID:15090507</ref>. See [[Molecular Playground/CheA]]. | |||
*'''CheB''' is a methyltransferase catalysing the demethylationof chemotaxis membrane receptors in ''Salmonella typhimurium''<ref>PMID:2991277</ref>. | |||
*'''CheW''' participates in the control of CheA activity<ref>PMID:1992467</ref>. See [[Molecular Playground/Bacterial Chemotaxis Receptors]] | |||
*'''PomA''' and '''PomB''' convert Na+ flux through channel to rotation of the flagellum in several bacteria <ref>PMID:21642461</ref>. | |||
*'''Aspartate chemotaxis receptor''' or '''Tar''' modulates the activity of CheA<ref>PMID:8832891</ref>. See [[DNA Origami Assembly for the Tar Chemoreceptor]]. | |||
*'''Methyl-accepting chemotaxis protein''' are the predominant chemoreceptors in bacteria and archaea. They are involved in regulation of diverse aspects of cellular activities<ref>PMID:28409190</ref>. See [[Molecular Playground/cytoplasmic domain of a serine chemotaxis receptor]]. | |||
*'''FLiN''', '''MotA''' and '''MotB''' are involved in energizing flagellar rotation in ''E. coli''<ref>PMID:7892209</ref>. | |||
*'''CKI1''' is a histidine kinase involved in cytokinin signal transduction<ref>PMID:8875940</ref>. See [[Receiver domain of sensor histidine kinase CKI1]]. | |||
See also: | |||
*[[Molecular Playground/Cytoplasmic domain of chemoreceptor of Thermotoga maritima]]<br /> | *[[Molecular Playground/Cytoplasmic domain of chemoreceptor of Thermotoga maritima]]<br /> | ||
==3D structures of chemotaxis protein == | ==3D structures of chemotaxis protein == | ||
[[Chemotaxis protein 3D structures]] | |||
</StructureSection> | |||
== References == | |||
<references/> | |||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Latest revision as of 11:28, 27 January 2025
Chemotaxis proteins direct cell movement according to certain chemicals in their environment.[1]
See also: 3D structures of chemotaxis proteinChemotaxis protein 3D structures
|
| ||||||||||
ReferencesReferences
- ↑ Adler J, Tso WW. "Decision"-making in bacteria: chemotactic response of Escherichia coli to conflicting stimuli. Science. 1974 Jun 21;184(4143):1292-4. PMID:4598187
- ↑ Sanna MG, Swanson RV, Bourret RB, Simon MI. Mutations in the chemotactic response regulator, CheY, that confer resistance to the phosphatase activity of CheZ. Mol Microbiol. 1995 Mar;15(6):1069-79. PMID:7623663 doi:10.1111/j.1365-2958.1995.tb02282.x
- ↑ Djordjevic S, Stock AM. Chemotaxis receptor recognition by protein methyltransferase CheR. Nat Struct Biol. 1998 Jun;5(6):446-50. PMID:9628482
- ↑ Jahreis K, Morrison TB, Garzón A, Parkinson JS. Chemotactic signaling by an Escherichia coli CheA mutant that lacks the binding domain for phosphoacceptor partners. J Bacteriol. 2004 May;186(9):2664-72. PMID:15090507 doi:10.1128/JB.186.9.2664-2672.2004
- ↑ Simms SA, Keane MG, Stock J. Multiple forms of the CheB methylesterase in bacterial chemosensing. J Biol Chem. 1985 Aug 25;260(18):10161-8. PMID:2991277
- ↑ Gegner JA, Dahlquist FW. Signal transduction in bacteria: CheW forms a reversible complex with the protein kinase CheA. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):750-4. PMID:1992467 doi:10.1073/pnas.88.3.750
- ↑ Yonekura K, Maki-Yonekura S, Homma M. Structure of the flagellar motor protein complex PomAB: implications for the torque-generating conformation. J Bacteriol. 2011 Aug;193(15):3863-70. PMID:21642461 doi:10.1128/JB.05021-11
- ↑ Tatsuno I, Homma M, Oosawa K, Kawagishi I. Signaling by the Escherichia coli aspartate chemoreceptor Tar with a single cytoplasmic domain per dimer. Science. 1996 Oct 18;274(5286):423-5. PMID:8832891 doi:10.1126/science.274.5286.423
- ↑ Salah Ud-Din AIM, Roujeinikova A. Methyl-accepting chemotaxis proteins: a core sensing element in prokaryotes and archaea. Cell Mol Life Sci. 2017 Sep;74(18):3293-3303. PMID:28409190 doi:10.1007/s00018-017-2514-0
- ↑ Garza AG, Harris-Haller LW, Stoebner RA, Manson MD. Motility protein interactions in the bacterial flagellar motor. Proc Natl Acad Sci U S A. 1995 Mar 14;92(6):1970-4. PMID:7892209 doi:10.1073/pnas.92.6.1970
- ↑ Kakimoto T. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science. 1996 Nov 8;274(5289):982-5. PMID:8875940