Replication Termination Protein: Difference between revisions
No edit summary |
Michal Harel (talk | contribs) No edit summary |
||
| (20 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
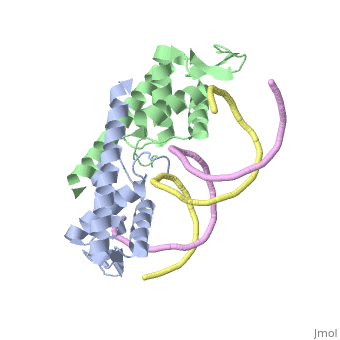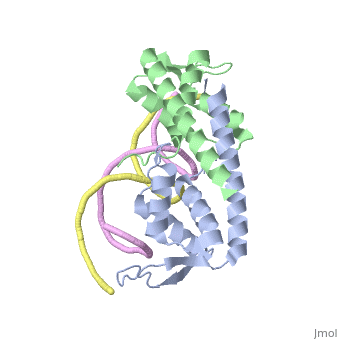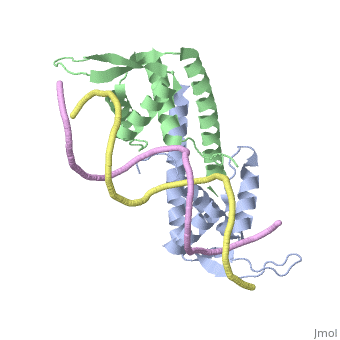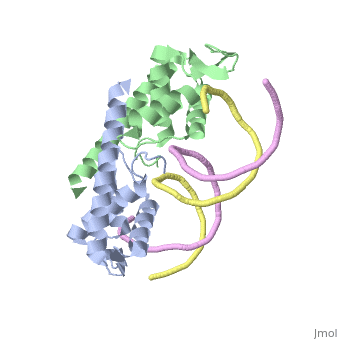
<StructureSection load=' | <StructureSection load='2dpd' size='450' side='right' scene='' caption='Assymetric RTP dimer bound to the B site in a Ter region (PDB code [[2dpd]])'> | ||
== Function == | == Function == | ||
The '''replication termination protein''' (RTP) is one of only two well-defined proteins known to be involved in arresting DNA replication forks, the other being a protein known as | [[Image:RTP.png|200px|left|thumb| Diagram of RTP monomer with secondary structure highlighted.]] | ||
The '''replication termination protein''' or '''replication terminator protein''' (RTP) is one of only two well-defined proteins known to be involved in arresting DNA replication forks, the other being a protein known as '''Tus''' ('''termination utilisation substance''' or '''DNA replication terminus site-binding protein''' or '''ter-binding protein)''' from ''E. coli'' <ref> Kamada K, Horiuchi T, Ohsumi K, Shimamoto N, Morkikawa K, (1996) Structure of a replication-terminator protein complexed with DNA. Nature, 383:598-603 </ref>. RTP was discovered in ''Bacillus subtilis'' and has been identified as a DNA binding protein of the winged helix family that forms a dimer of 29kDa. This dimeric form has been shown to have an exceptionally high affinity for its cognate binding sites( Kd ~10-11M-1)<ref> Wilce et. al. (2001) Structure of the RTP−DNA complex and the mechanism of polar replication fork arrest. Nature Structural Biology, 8:206-210 </ref>, otherwise known as Termination sites (Ter sites). These Ter sites are found in multiple locations in the ''B. subtilis'' genome <ref> Gautam A. et.al. (2001) A single domain of the replication termination protein of ''Bacillus subtilis'' is involved in arresting both DnaB helicase and RNA polymerase. Journal of Biological Chemistry, 276:23471-23479</ref>. For more details see:<br /> | |||
*[[RTP and Tus]]<br /> | *[[RTP and Tus]]<br /> | ||
*[[Rtp and Tus DNA Binding]]<br /> | *[[Rtp and Tus DNA Binding]]<br /> | ||
| Line 20: | Line 19: | ||
[[Image:yehhhh.png|350px|left|thumb| B. subtilis genome showing origin of replication, polar Ter sites and both replication forks.]] | [[Image:yehhhh.png|350px|left|thumb| B. subtilis genome showing origin of replication, polar Ter sites and both replication forks.]] | ||
{{Clear}} | {{Clear}} | ||
Bacterial replication like that found in Bacillus subtilis consists of two replication forks that travel in opposite directions around the same circular strand of DNA. These replication forks begin at the origin of replication (OriC) and travel in clockwise and anticlockwise directions <ref> Noirot P (2007). "Replication of the Bacillus subtilis chromosome". In Graumann P. Bacillus: Cellular and Molecular Biology. Caister Academic Press. ISBN 978-1-904455-12-7 </ref>. To prevent the strand from being over replicated each strand needs to be terminated roughly opposite the OriC and then joined to form the complete duplicated strand of DNA. As such the Ter/RTP complexes have found to be polar in their action; that is that different Ter sites are capable of only blacking DNA replication from one direction only. Since Ter sites are found facing both forks of replication, and the combination of both are capable of stopping both strands of replication, they are often termed as replication traps. | Bacterial replication like that found in ''Bacillus subtilis'' consists of two replication forks that travel in opposite directions around the same circular strand of DNA. These replication forks begin at the origin of replication (OriC) and travel in clockwise and anticlockwise directions <ref> Noirot P (2007). "Replication of the Bacillus subtilis chromosome". In Graumann P. Bacillus: Cellular and Molecular Biology. Caister Academic Press. ISBN 978-1-904455-12-7 </ref>. To prevent the strand from being over replicated each strand needs to be terminated roughly opposite the OriC and then joined to form the complete duplicated strand of DNA. As such the Ter/RTP complexes have found to be polar in their action; that is that different Ter sites are capable of only blacking DNA replication from one direction only. Since Ter sites are found facing both forks of replication, and the combination of both are capable of stopping both strands of replication, they are often termed as replication traps. | ||
The arresting of replication is achieved by the binding of two dimers to each Ter site, with the two RTP binding sites referred to as site A and site B. Site B shows a higher affinity for RTP and binds first, with site A then being filled with another RTP dimer cooperatively. This differential affinity for RTP is cited as a possible reason for the observed polarity of the termination <ref> Duggin I.G. (2006) DNA Replication Fork Arrest by the Bacillus subtilis RTP–DNA Complex Involves a Mechanism that Is Independent of the Affinity of RTP–DNA Binding. Journal of Molecular Biology, 361:1-6 </ref>. | The arresting of replication is achieved by the binding of two dimers to each Ter site, with the two RTP binding sites referred to as site A and site B. Site B shows a higher affinity for RTP and binds first, with site A then being filled with another RTP dimer cooperatively. This differential affinity for RTP is cited as a possible reason for the observed polarity of the termination <ref> Duggin I.G. (2006) DNA Replication Fork Arrest by the ''Bacillus subtilis'' RTP–DNA Complex Involves a Mechanism that Is Independent of the Affinity of RTP–DNA Binding. Journal of Molecular Biology, 361:1-6 </ref>. | ||
==RTP Structure== | ==RTP Structure== | ||
RTP is a DNA binding protein from Bacillus Subtilis that uses a helix-loop-helix binding motif. In solution it shows a symmetric structure typical of the winged helix loop helix family, with an unstructured <scene name='44/449701/1/3'>N-terminus</scene> end, first alpha helix <scene name='44/449701/1/4'>(a1)</scene>, unstructured loop that is equivalent to the first beta sheet <scene name=' | RTP is a DNA binding protein from Bacillus Subtilis that uses a helix-loop-helix binding motif. In solution it shows a symmetric structure typical of the winged helix loop helix family, with an unstructured <scene name='44/449701/1/3'>N-terminus</scene> end, first alpha helix <scene name='44/449701/1/4'>(a1)</scene>, unstructured loop that is equivalent to the first beta sheet <scene name='44/449701/1/6'>(B1)</scene>, helix loop helix structure (<scene name='44/449701/1/5'>a2</scene>-<scene name='44/449701/1/7'>a3</scene>), 2 beta sheets with a connecting loop that makes up the 'wing' structure <scene name='44/449701/1/8'>(B2 - B3)</scene> and an additional long alpha helix involved in dimerisation <scene name='44/449701/1/9'>(a4)</scene>.<ref>Vivian JP, Porter CJ, Wilce JA, Wilce MCJ, (2007) An asymmetric structure of the Bacillus subtilise Replication Terminator Protein in Complex with DNA. J. Mol. Bio, 370:481-491</ref> | ||
Binding of Rtp to the assymetric B portion of the Ter site changes its conformation into an assymetric 'wing-up wing-down' structure. | Binding of Rtp to the assymetric B portion of the Ter site changes its conformation into an assymetric 'wing-up wing-down' structure. | ||
The <scene name=' | The <scene name='44/449701/1/10'>wing-up</scene> contacts upstream with rtp dimer bound to A-site while the <scene name='44/449701/1/11'>wing-down</scene>: Contacts with phosphate backbone of downstream DNA | ||
| Line 120: | Line 119: | ||
Debate over the mechanism of Replication termination by RTP has led to a number of mechanisms being proposed. Original work on this question was hampered by an incorrect structure of RTP being released. This structure was produced in solution and was shown to be a symmetric dimer<ref> Bastia D. (1995) Crystal structure of the replication terminator protein from b. subtilis at 2.6 A. Cell 80: 651-660 </ref> Later crystallisation of the protein bound to DNA showed the RTP formed a asymmetric dimer when bound to cognate sequences <ref> Wilce et. al. (2001) Structure of the RTP−DNA complex and the mechanism of polar replication fork arrest. Nature Structural Biology, 8:206-210</ref> . One early study proposed that the asymmetric interaction between terminator protein and terminator DNA contributed to the observed polarity, with later studies showing that the RTP/Ter complex contained partially unwound DNA, suggesting a locked complex was responsible. More recent data has implicated that protein -protein interactions between RTP and the helicase is primarily responsible for termination and that this mechanism is modulated by the asymmetrical interactions described above <ref>Kaplan D.L., Bastia D. (2009). Mechanisms of polar arrest of a replication fork. Molecular Microbiology 72: 279-284</ref>. | Debate over the mechanism of Replication termination by RTP has led to a number of mechanisms being proposed. Original work on this question was hampered by an incorrect structure of RTP being released. This structure was produced in solution and was shown to be a symmetric dimer<ref> Bastia D. (1995) Crystal structure of the replication terminator protein from b. subtilis at 2.6 A. Cell 80: 651-660 </ref> Later crystallisation of the protein bound to DNA showed the RTP formed a asymmetric dimer when bound to cognate sequences <ref> Wilce et. al. (2001) Structure of the RTP−DNA complex and the mechanism of polar replication fork arrest. Nature Structural Biology, 8:206-210</ref> . One early study proposed that the asymmetric interaction between terminator protein and terminator DNA contributed to the observed polarity, with later studies showing that the RTP/Ter complex contained partially unwound DNA, suggesting a locked complex was responsible. More recent data has implicated that protein -protein interactions between RTP and the helicase is primarily responsible for termination and that this mechanism is modulated by the asymmetrical interactions described above <ref>Kaplan D.L., Bastia D. (2009). Mechanisms of polar arrest of a replication fork. Molecular Microbiology 72: 279-284</ref>. | ||
==3D structures of replication termination protein== | |||
[[Replication termination protein 3D structures]] | |||
[[ | |||
==References== | ==References== | ||
{{reflist}} | {{reflist}} | ||
</StructureSection> | |||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Latest revision as of 11:36, 6 August 2024
Function The replication termination protein or replication terminator protein (RTP) is one of only two well-defined proteins known to be involved in arresting DNA replication forks, the other being a protein known as Tus (termination utilisation substance or DNA replication terminus site-binding protein or ter-binding protein) from E. coli [1]. RTP was discovered in Bacillus subtilis and has been identified as a DNA binding protein of the winged helix family that forms a dimer of 29kDa. This dimeric form has been shown to have an exceptionally high affinity for its cognate binding sites( Kd ~10-11M-1)[2], otherwise known as Termination sites (Ter sites). These Ter sites are found in multiple locations in the B. subtilis genome [3]. For more details see:
Termination of Replication Bacterial replication like that found in Bacillus subtilis consists of two replication forks that travel in opposite directions around the same circular strand of DNA. These replication forks begin at the origin of replication (OriC) and travel in clockwise and anticlockwise directions [4]. To prevent the strand from being over replicated each strand needs to be terminated roughly opposite the OriC and then joined to form the complete duplicated strand of DNA. As such the Ter/RTP complexes have found to be polar in their action; that is that different Ter sites are capable of only blacking DNA replication from one direction only. Since Ter sites are found facing both forks of replication, and the combination of both are capable of stopping both strands of replication, they are often termed as replication traps. The arresting of replication is achieved by the binding of two dimers to each Ter site, with the two RTP binding sites referred to as site A and site B. Site B shows a higher affinity for RTP and binds first, with site A then being filled with another RTP dimer cooperatively. This differential affinity for RTP is cited as a possible reason for the observed polarity of the termination [5]. RTP StructureRTP is a DNA binding protein from Bacillus Subtilis that uses a helix-loop-helix binding motif. In solution it shows a symmetric structure typical of the winged helix loop helix family, with an unstructured end, first alpha helix , unstructured loop that is equivalent to the first beta sheet , helix loop helix structure (-), 2 beta sheets with a connecting loop that makes up the 'wing' structure and an additional long alpha helix involved in dimerisation .[6] Binding of Rtp to the assymetric B portion of the Ter site changes its conformation into an assymetric 'wing-up wing-down' structure. The contacts upstream with rtp dimer bound to A-site while the : Contacts with phosphate backbone of downstream DNA
Residues that bind in both monomers: Red Residues that bind only in wing-up: Green Residues that bind only in wing-down: Blue
Replication MechanismDebate over the mechanism of Replication termination by RTP has led to a number of mechanisms being proposed. Original work on this question was hampered by an incorrect structure of RTP being released. This structure was produced in solution and was shown to be a symmetric dimer[8] Later crystallisation of the protein bound to DNA showed the RTP formed a asymmetric dimer when bound to cognate sequences [9] . One early study proposed that the asymmetric interaction between terminator protein and terminator DNA contributed to the observed polarity, with later studies showing that the RTP/Ter complex contained partially unwound DNA, suggesting a locked complex was responsible. More recent data has implicated that protein -protein interactions between RTP and the helicase is primarily responsible for termination and that this mechanism is modulated by the asymmetrical interactions described above [10]. 3D structures of replication termination proteinReplication termination protein 3D structures References
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||