3frp: Difference between revisions
No edit summary |
No edit summary |
||
| (12 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
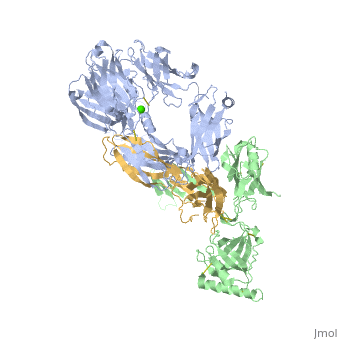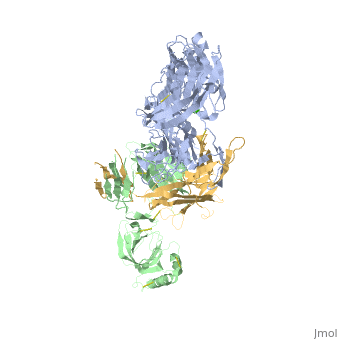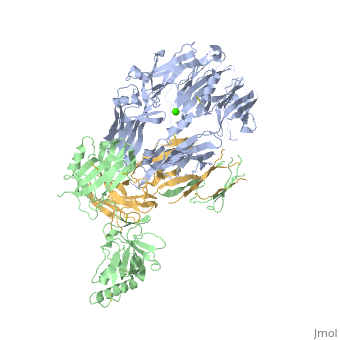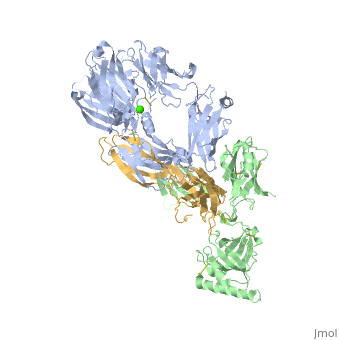
==Crystal Structure of Cobra Venom Factor, a Co-factor for C3- and C5 convertase CVFBb== | |||
<StructureSection load='3frp' size='340' side='right'caption='[[3frp]], [[Resolution|resolution]] 2.61Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[3frp]] is a 3 chain structure with sequence from [https://en.wikipedia.org/wiki/Naja_kaouthia Naja kaouthia]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3FRP OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3FRP FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.61Å</td></tr> | |||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=CA:CALCIUM+ION'>CA</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3frp FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3frp OCA], [https://pdbe.org/3frp PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3frp RCSB], [https://www.ebi.ac.uk/pdbsum/3frp PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3frp ProSAT]</span></td></tr> | |||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/VCO3_NAJKA VCO3_NAJKA] Complement-activating protein in cobra venom. It is a structural and functional analog of complement component C3b, the activated form of C3. It binds factor B (CFB), which is subsequently cleaved by factor D (CFD) to form the bimolecular complex CVF/Bb. CVF/Bb is a C3/C5 convertase that cleaves both complement components C3 and C5. Structurally, it resembles the C3b degradation product C3c, which is not able to form a C3/C5 convertase. Unlike C3b/Bb, CVF/Bb is a stable complex and completely resistant to the actions of complement regulatory factors H (CFH) and I (CFI). Therefore, CVF continuously activates complement resulting in the depletion of complement activity. | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/fr/3frp_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=3frp ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
Cobra venom factor (CVF) is a functional analog of human complement component C3b, the active fragment of C3. Similar to C3b, in human and mammalian serum, CVF binds factor B, which is then cleaved by factor D, giving rise to the CVFBb complex that targets the same scissile bond in C3 as the authentic complement convertases C4bC2a and C3bBb. Unlike the latter, CVFBb is a stable complex and an efficient C5 convertase. We solved the crystal structure of CVF, isolated from Naja naja kouthia venom, at 2.6 A resolution. The CVF crystal structure, an intermediate between C3b and C3c, lacks the TED domain and has the CUB domain in an identical position to that seen in C3b. The similarly positioned CUB and slightly displaced C345c domains of CVF could play a vital role in the formation of C3 convertases by providing important primary binding sites for factor B. | |||
The crystal structure of cobra venom factor, a cofactor for C3- and C5-convertase CVFBb.,Krishnan V, Ponnuraj K, Xu Y, Macon K, Volanakis JE, Narayana SV Structure. 2009 Apr 15;17(4):611-9. PMID:19368894<ref>PMID:19368894</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 3frp" style="background-color:#fffaf0;"></div> | |||
==See Also== | |||
*[[Cobra venom factor|Cobra venom factor]] | |||
== References == | |||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
[[Category: Large Structures]] | |||
[[Category: Naja kaouthia]] | |||
[[Category: Krishnan V]] | |||
[[Category: Narayana SVL]] | |||
Latest revision as of 04:49, 21 November 2024
Crystal Structure of Cobra Venom Factor, a Co-factor for C3- and C5 convertase CVFBbCrystal Structure of Cobra Venom Factor, a Co-factor for C3- and C5 convertase CVFBb
Structural highlights
FunctionVCO3_NAJKA Complement-activating protein in cobra venom. It is a structural and functional analog of complement component C3b, the activated form of C3. It binds factor B (CFB), which is subsequently cleaved by factor D (CFD) to form the bimolecular complex CVF/Bb. CVF/Bb is a C3/C5 convertase that cleaves both complement components C3 and C5. Structurally, it resembles the C3b degradation product C3c, which is not able to form a C3/C5 convertase. Unlike C3b/Bb, CVF/Bb is a stable complex and completely resistant to the actions of complement regulatory factors H (CFH) and I (CFI). Therefore, CVF continuously activates complement resulting in the depletion of complement activity. Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedCobra venom factor (CVF) is a functional analog of human complement component C3b, the active fragment of C3. Similar to C3b, in human and mammalian serum, CVF binds factor B, which is then cleaved by factor D, giving rise to the CVFBb complex that targets the same scissile bond in C3 as the authentic complement convertases C4bC2a and C3bBb. Unlike the latter, CVFBb is a stable complex and an efficient C5 convertase. We solved the crystal structure of CVF, isolated from Naja naja kouthia venom, at 2.6 A resolution. The CVF crystal structure, an intermediate between C3b and C3c, lacks the TED domain and has the CUB domain in an identical position to that seen in C3b. The similarly positioned CUB and slightly displaced C345c domains of CVF could play a vital role in the formation of C3 convertases by providing important primary binding sites for factor B. The crystal structure of cobra venom factor, a cofactor for C3- and C5-convertase CVFBb.,Krishnan V, Ponnuraj K, Xu Y, Macon K, Volanakis JE, Narayana SV Structure. 2009 Apr 15;17(4):611-9. PMID:19368894[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||