2pfb: Difference between revisions
No edit summary |
No edit summary |
||
| (11 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
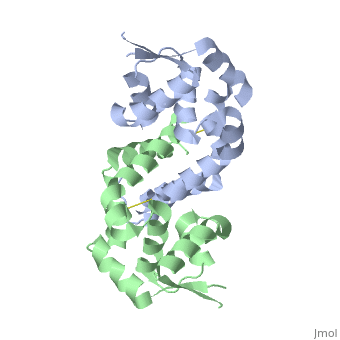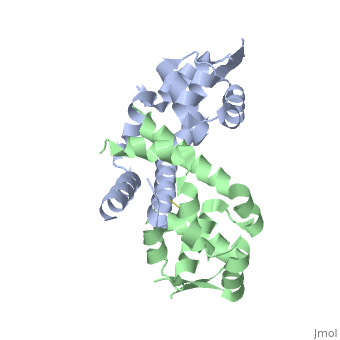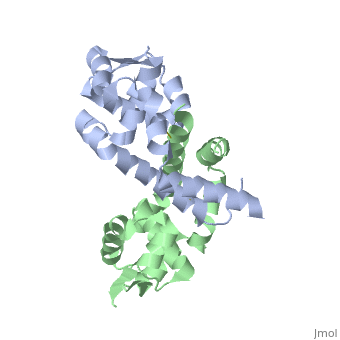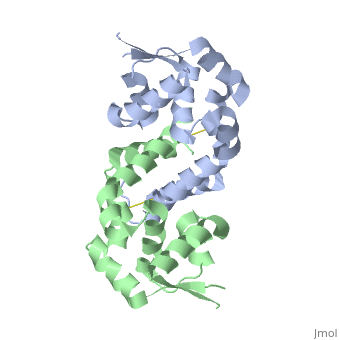
< | ==Structure of oxidized OhrR from Xanthamonas campestris== | ||
<StructureSection load='2pfb' size='340' side='right'caption='[[2pfb]], [[Resolution|resolution]] 1.93Å' scene=''> | |||
You may | == Structural highlights == | ||
<table><tr><td colspan='2'>[[2pfb]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Xanthomonas_campestris Xanthomonas campestris]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2PFB OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2PFB FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.93Å</td></tr> | |||
- | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2pfb FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2pfb OCA], [https://pdbe.org/2pfb PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2pfb RCSB], [https://www.ebi.ac.uk/pdbsum/2pfb PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2pfb ProSAT]</span></td></tr> | ||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/Q93R11_XANCH Q93R11_XANCH] | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/pf/2pfb_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=2pfb ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
The Xanthomonas campestris transcription regulator OhrR contains a reactive cysteine residue (C22) that upon oxidation by organic hydroperoxides (OHPs) forms an intersubunit disulphide bond with residue C127'. Such modification induces the expression of a peroxidase that reduces OHPs to their less toxic alcohols. Here, we describe the structures of reduced and OHP-oxidized OhrR, visualizing the structural mechanism of OHP induction. Reduced OhrR takes a canonical MarR family fold with C22 and C127' separated by 15.5 A. OHP oxidation results in the disruption of the Y36'-C22-Y47' interaction network and dissection of helix alpha5, which then allows the 135 degrees rotation and 8.2 A translation of C127', formation of the C22-C127' disulphide bond, and alpha6-alpha6' helix-swapped reconfiguration of the dimer interface. These changes result in the 28 degrees rigid body rotations of each winged helix-turn-helix motif and DNA dissociation. Similar effector-induced rigid body rotations are expected for most MarR family members. | |||
Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR.,Newberry KJ, Fuangthong M, Panmanee W, Mongkolsuk S, Brennan RG Mol Cell. 2007 Nov 30;28(4):652-64. PMID:18042459<ref>PMID:18042459</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 2pfb" style="background-color:#fffaf0;"></div> | |||
==See Also== | |||
*[[Organic hydroperoxide resistance protein|Organic hydroperoxide resistance protein]] | |||
== References == | |||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
== | [[Category: Large Structures]] | ||
== | |||
< | |||
[[Category: Xanthomonas campestris]] | [[Category: Xanthomonas campestris]] | ||
[[Category: Brennan | [[Category: Brennan RG]] | ||
[[Category: Newberry | [[Category: Newberry KJ]] | ||
Latest revision as of 11:28, 30 October 2024
Structure of oxidized OhrR from Xanthamonas campestrisStructure of oxidized OhrR from Xanthamonas campestris
Structural highlights
FunctionEvolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedThe Xanthomonas campestris transcription regulator OhrR contains a reactive cysteine residue (C22) that upon oxidation by organic hydroperoxides (OHPs) forms an intersubunit disulphide bond with residue C127'. Such modification induces the expression of a peroxidase that reduces OHPs to their less toxic alcohols. Here, we describe the structures of reduced and OHP-oxidized OhrR, visualizing the structural mechanism of OHP induction. Reduced OhrR takes a canonical MarR family fold with C22 and C127' separated by 15.5 A. OHP oxidation results in the disruption of the Y36'-C22-Y47' interaction network and dissection of helix alpha5, which then allows the 135 degrees rotation and 8.2 A translation of C127', formation of the C22-C127' disulphide bond, and alpha6-alpha6' helix-swapped reconfiguration of the dimer interface. These changes result in the 28 degrees rigid body rotations of each winged helix-turn-helix motif and DNA dissociation. Similar effector-induced rigid body rotations are expected for most MarR family members. Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR.,Newberry KJ, Fuangthong M, Panmanee W, Mongkolsuk S, Brennan RG Mol Cell. 2007 Nov 30;28(4):652-64. PMID:18042459[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||