1jb0: Difference between revisions
No edit summary |
No edit summary |
||
| (5 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
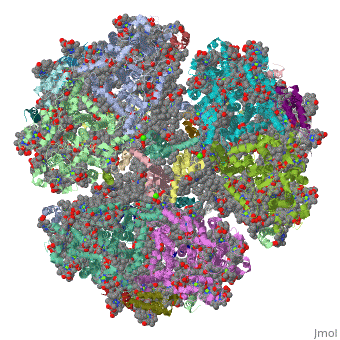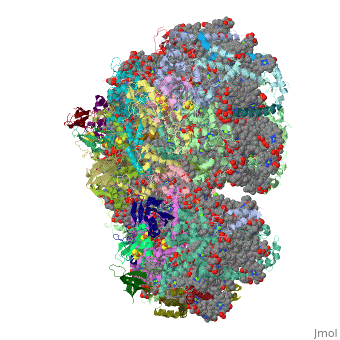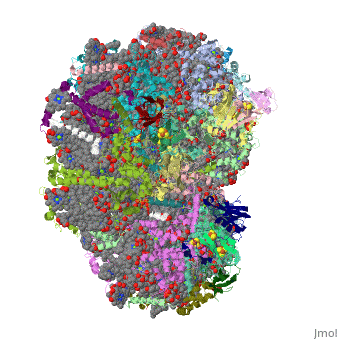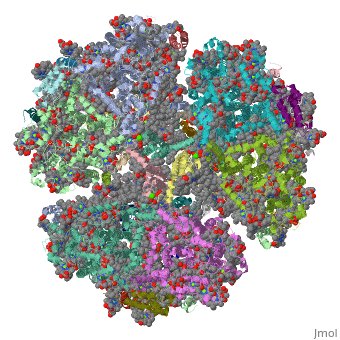
==Crystal Structure of Photosystem I: a Photosynthetic Reaction Center and Core Antenna System from Cyanobacteria== | ==Crystal Structure of Photosystem I: a Photosynthetic Reaction Center and Core Antenna System from Cyanobacteria== | ||
<StructureSection load='1jb0' size='340' side='right' caption='[[1jb0]], [[Resolution|resolution]] 2.50Å' scene=''> | <StructureSection load='1jb0' size='340' side='right'caption='[[1jb0]], [[Resolution|resolution]] 2.50Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[1jb0]] is a | <table><tr><td colspan='2'>[[1jb0]] is a 10 chain structure with sequence from [https://en.wikipedia.org/wiki/Synechococcus_elongatus Synechococcus elongatus]. The October 2001 RCSB PDB [https://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''Photosystem I'' by David S. Goodsell is [https://dx.doi.org/10.2210/rcsb_pdb/mom_2001_10 10.2210/rcsb_pdb/mom_2001_10]. The June 2005 RCSB PDB [https://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''Carotenoid Oxygenase'' by David S. Goodsell is [https://dx.doi.org/10.2210/rcsb_pdb/mom_2005_6 10.2210/rcsb_pdb/mom_2005_6]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1JB0 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1JB0 FirstGlance]. <br> | ||
</td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=BCR:BETA-CAROTENE'>BCR</scene>, <scene name='pdbligand=CA:CALCIUM+ION'>CA</scene>, <scene name='pdbligand=CLA:CHLOROPHYLL+A'>CLA</scene>, <scene name='pdbligand=LHG:1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE'>LHG</scene>, <scene name='pdbligand=LMG:1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE'>LMG</scene>, <scene name='pdbligand=PQN:PHYLLOQUINONE'>PQN | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.5Å</td></tr> | ||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=BCR:BETA-CAROTENE'>BCR</scene>, <scene name='pdbligand=CA:CALCIUM+ION'>CA</scene>, <scene name='pdbligand=CLA:CHLOROPHYLL+A'>CLA</scene>, <scene name='pdbligand=LHG:1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE'>LHG</scene>, <scene name='pdbligand=LMG:1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE'>LMG</scene>, <scene name='pdbligand=PQN:PHYLLOQUINONE'>PQN</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1jb0 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1jb0 OCA], [https://pdbe.org/1jb0 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1jb0 RCSB], [https://www.ebi.ac.uk/pdbsum/1jb0 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1jb0 ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
[ | [https://www.uniprot.org/uniprot/PSAA_THEVB PSAA_THEVB] PsaA and PsaB bind P700, the primary electron donor of photosystem I (PSI), as well as the electron acceptors A0, A1 and FX. PSI is a plastocyanin/cytochrome c6-ferredoxin oxidoreductase, converting photonic excitation into a charge separation, which transfers an electron from the donor P700 chlorophyll pair to the spectroscopically characterized acceptors A0, A1, FX, FA and FB in turn. Oxidized P700 is reduced on the lumenal side of the thylakoid membrane by plastocyanin or cytochrome c6.<ref>PMID:10066799</ref> <ref>PMID:10066800</ref> <ref>PMID:11418848</ref> <ref>PMID:8901876</ref> | ||
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
Check<jmol> | Check<jmol> | ||
<jmolCheckbox> | <jmolCheckbox> | ||
<scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/jb/1jb0_consurf.spt"</scriptWhenChecked> | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/jb/1jb0_consurf.spt"</scriptWhenChecked> | ||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/ | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> | ||
<text>to colour the structure by Evolutionary Conservation</text> | <text>to colour the structure by Evolutionary Conservation</text> | ||
</jmolCheckbox> | </jmolCheckbox> | ||
| Line 30: | Line 31: | ||
==See Also== | ==See Also== | ||
*[[Photosystem I|Photosystem I]] | *[[Photosystem I|Photosystem I]] | ||
*[[Photosystem I | *[[Photosystem I 3D structures|Photosystem I 3D structures]] | ||
== References == | == References == | ||
<references/> | <references/> | ||
| Line 39: | Line 38: | ||
</StructureSection> | </StructureSection> | ||
[[Category: Carotenoid Oxygenase]] | [[Category: Carotenoid Oxygenase]] | ||
[[Category: Large Structures]] | |||
[[Category: Photosystem I]] | [[Category: Photosystem I]] | ||
[[Category: RCSB PDB Molecule of the Month]] | [[Category: RCSB PDB Molecule of the Month]] | ||
[[Category: Synechococcus elongatus]] | [[Category: Synechococcus elongatus]] | ||
[[Category: Fromme | [[Category: Fromme P]] | ||
[[Category: Jordan | [[Category: Jordan P]] | ||
[[Category: Klukas | [[Category: Klukas O]] | ||
[[Category: Krauss | [[Category: Krauss N]] | ||
[[Category: Saenger | [[Category: Saenger W]] | ||
[[Category: Witt | [[Category: Witt HT]] | ||
Latest revision as of 09:48, 30 October 2024
Crystal Structure of Photosystem I: a Photosynthetic Reaction Center and Core Antenna System from CyanobacteriaCrystal Structure of Photosystem I: a Photosynthetic Reaction Center and Core Antenna System from Cyanobacteria
Structural highlights
FunctionPSAA_THEVB PsaA and PsaB bind P700, the primary electron donor of photosystem I (PSI), as well as the electron acceptors A0, A1 and FX. PSI is a plastocyanin/cytochrome c6-ferredoxin oxidoreductase, converting photonic excitation into a charge separation, which transfers an electron from the donor P700 chlorophyll pair to the spectroscopically characterized acceptors A0, A1, FX, FA and FB in turn. Oxidized P700 is reduced on the lumenal side of the thylakoid membrane by plastocyanin or cytochrome c6.[1] [2] [3] [4] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedLife on Earth depends on photosynthesis, the conversion of light energy from the Sun to chemical energy. In plants, green algae and cyanobacteria, this process is driven by the cooperation of two large protein-cofactor complexes, photosystems I and II, which are located in the thylakoid photosynthetic membranes. The crystal structure of photosystem I from the thermophilic cyanobacterium Synechococcus elongatus described here provides a picture at atomic detail of 12 protein subunits and 127 cofactors comprising 96 chlorophylls, 2 phylloquinones, 3 Fe4S4 clusters, 22 carotenoids, 4 lipids, a putative Ca2+ ion and 201 water molecules. The structural information on the proteins and cofactors and their interactions provides a basis for understanding how the high efficiency of photosystem I in light capturing and electron transfer is achieved. Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution.,Jordan P, Fromme P, Witt HT, Klukas O, Saenger W, Krauss N Nature. 2001 Jun 21;411(6840):909-17. PMID:11418848[5] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||