2qc8: Difference between revisions
No edit summary |
No edit summary |
||
| (18 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
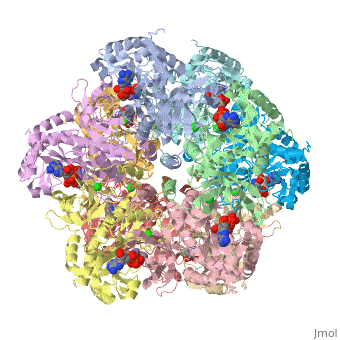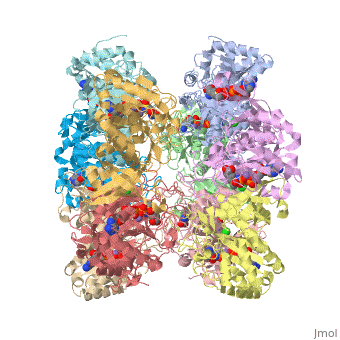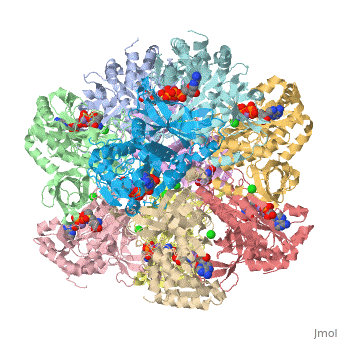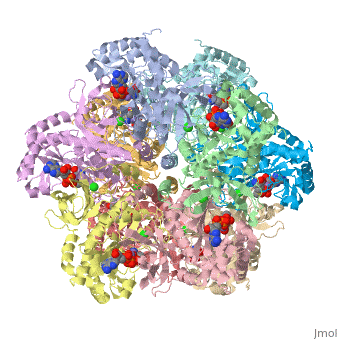
== | ==Crystal structure of human glutamine synthetase in complex with ADP and methionine sulfoximine phosphate== | ||
Glutamine synthetase (GS) catalyzes the ligation of glutamate and ammonia | <StructureSection load='2qc8' size='340' side='right'caption='[[2qc8]], [[Resolution|resolution]] 2.60Å' scene=''> | ||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[2qc8]] is a 10 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2QC8 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2QC8 FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.6Å</td></tr> | |||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=ADP:ADENOSINE-5-DIPHOSPHATE'>ADP</scene>, <scene name='pdbligand=CL:CHLORIDE+ION'>CL</scene>, <scene name='pdbligand=MN:MANGANESE+(II)+ION'>MN</scene>, <scene name='pdbligand=P3S:L-METHIONINE-S-SULFOXIMINE+PHOSPHATE'>P3S</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2qc8 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2qc8 OCA], [https://pdbe.org/2qc8 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2qc8 RCSB], [https://www.ebi.ac.uk/pdbsum/2qc8 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2qc8 ProSAT]</span></td></tr> | |||
</table> | |||
== Disease == | |||
[https://www.uniprot.org/uniprot/GLNA_HUMAN GLNA_HUMAN] Defects in GLUL are the cause of congenital systemic glutamine deficiency (CSGD) [MIM:[https://omim.org/entry/610015 610015]. CSGD is a rare developmental disorder with severe brain malformation resulting in multi-organ failure and neonatal death. Glutamine is largely absent from affected patients serum, urine and cerebrospinal fluid.<ref>PMID:16267323</ref> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/GLNA_HUMAN GLNA_HUMAN] This enzyme has 2 functions: it catalyzes the production of glutamine and 4-aminobutanoate (gamma-aminobutyric acid, GABA), the latter in a pyridoxal phosphate-independent manner (By similarity). Essential for proliferation of fetal skin fibroblasts.<ref>PMID:18662667</ref> | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/qc/2qc8_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=2qc8 ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
Glutamine synthetase (GS) catalyzes the ligation of glutamate and ammonia to form glutamine, with concomitant hydrolysis of ATP. In mammals, the activity eliminates cytotoxic ammonia, at the same time converting neurotoxic glutamate to harmless glutamine; there are a number of links between changes in GS activity and neurodegenerative disorders, such as Alzheimer's disease. In plants, because of its importance in the assimilation and re-assimilation of ammonia, the enzyme is a target of some herbicides. GS is also a central component of bacterial nitrogen metabolism and a potential drug target. Previous studies had investigated the structures of bacterial and plant GSs. In the present publication, we report the first structures of mammalian GSs. The apo form of the canine enzyme was solved by molecular replacement and refined at a resolution of 3 A. Two structures of human glutamine synthetase represent complexes with: a) phosphate, ADP, and manganese, and b) a phosphorylated form of the inhibitor methionine sulfoximine, ADP and manganese; these structures were refined to resolutions of 2.05 A and 2.6 A, respectively. Loop movements near the active site generate more closed forms of the eukaryotic enzymes when substrates are bound; the largest changes are associated with the binding of the nucleotide. Comparisons with earlier structures provide a basis for the design of drugs that are specifically directed at either human or bacterial enzymes. The site of binding the amino acid substrate is highly conserved in bacterial and eukaryotic GSs, whereas the nucleotide binding site varies to a much larger degree. Thus, the latter site offers the best target for specific drug design. Differences between mammalian and plant enzymes are much more subtle, suggesting that herbicides targeting GS must be designed with caution. | |||
Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design.,Krajewski WW, Collins R, Holmberg-Schiavone L, Jones TA, Karlberg T, Mowbray SL J Mol Biol. 2008 Jan 4;375(1):217-28. Epub 2007 Oct 17. PMID:18005987<ref>PMID:18005987</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
[[ | <div class="pdbe-citations 2qc8" style="background-color:#fffaf0;"></div> | ||
==See Also== | |||
*[[Glutamine synthetase 3D structures|Glutamine synthetase 3D structures]] | |||
== References == | |||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
[[Category: | [[Category: Large Structures]] | ||
[[Category: Arrowsmith | [[Category: Arrowsmith CH]] | ||
[[Category: Berglund H]] | |||
[[Category: Berglund | [[Category: Busam RD]] | ||
[[Category: Busam | [[Category: Collins R]] | ||
[[Category: Collins | [[Category: Dahlgren LG]] | ||
[[Category: Dahlgren | [[Category: Edwards A]] | ||
[[Category: Edwards | [[Category: Flodin S]] | ||
[[Category: Flodin | [[Category: Flores A]] | ||
[[Category: Flores | [[Category: Graslund S]] | ||
[[Category: Graslund | [[Category: Hammarstrom M]] | ||
[[Category: Hammarstrom | [[Category: Hogbom M]] | ||
[[Category: Hogbom | [[Category: Holmberg-Schiavone L]] | ||
[[Category: Holmberg-Schiavone | [[Category: Johansson I]] | ||
[[Category: Johansson | [[Category: Kallas A]] | ||
[[Category: Kallas | [[Category: Karlberg T]] | ||
[[Category: Karlberg | [[Category: Kotenyova T]] | ||
[[Category: Kotenyova | [[Category: Lehtio L]] | ||
[[Category: Lehtio | [[Category: Moche M]] | ||
[[Category: Moche | [[Category: Nordlund P]] | ||
[[Category: Nordlund | [[Category: Nyman T]] | ||
[[Category: Nyman | [[Category: Persson C]] | ||
[[Category: Persson | [[Category: Sagemark J]] | ||
[[Category: Sundstrom M]] | |||
[[Category: Sagemark | [[Category: Thorsell AG]] | ||
[[Category: Sundstrom | [[Category: Van Den Berg S]] | ||
[[Category: Thorsell | [[Category: Weigelt J]] | ||
[[Category: | |||
[[Category: | |||
Latest revision as of 11:32, 30 October 2024
Crystal structure of human glutamine synthetase in complex with ADP and methionine sulfoximine phosphateCrystal structure of human glutamine synthetase in complex with ADP and methionine sulfoximine phosphate
Structural highlights
DiseaseGLNA_HUMAN Defects in GLUL are the cause of congenital systemic glutamine deficiency (CSGD) [MIM:610015. CSGD is a rare developmental disorder with severe brain malformation resulting in multi-organ failure and neonatal death. Glutamine is largely absent from affected patients serum, urine and cerebrospinal fluid.[1] FunctionGLNA_HUMAN This enzyme has 2 functions: it catalyzes the production of glutamine and 4-aminobutanoate (gamma-aminobutyric acid, GABA), the latter in a pyridoxal phosphate-independent manner (By similarity). Essential for proliferation of fetal skin fibroblasts.[2] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedGlutamine synthetase (GS) catalyzes the ligation of glutamate and ammonia to form glutamine, with concomitant hydrolysis of ATP. In mammals, the activity eliminates cytotoxic ammonia, at the same time converting neurotoxic glutamate to harmless glutamine; there are a number of links between changes in GS activity and neurodegenerative disorders, such as Alzheimer's disease. In plants, because of its importance in the assimilation and re-assimilation of ammonia, the enzyme is a target of some herbicides. GS is also a central component of bacterial nitrogen metabolism and a potential drug target. Previous studies had investigated the structures of bacterial and plant GSs. In the present publication, we report the first structures of mammalian GSs. The apo form of the canine enzyme was solved by molecular replacement and refined at a resolution of 3 A. Two structures of human glutamine synthetase represent complexes with: a) phosphate, ADP, and manganese, and b) a phosphorylated form of the inhibitor methionine sulfoximine, ADP and manganese; these structures were refined to resolutions of 2.05 A and 2.6 A, respectively. Loop movements near the active site generate more closed forms of the eukaryotic enzymes when substrates are bound; the largest changes are associated with the binding of the nucleotide. Comparisons with earlier structures provide a basis for the design of drugs that are specifically directed at either human or bacterial enzymes. The site of binding the amino acid substrate is highly conserved in bacterial and eukaryotic GSs, whereas the nucleotide binding site varies to a much larger degree. Thus, the latter site offers the best target for specific drug design. Differences between mammalian and plant enzymes are much more subtle, suggesting that herbicides targeting GS must be designed with caution. Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design.,Krajewski WW, Collins R, Holmberg-Schiavone L, Jones TA, Karlberg T, Mowbray SL J Mol Biol. 2008 Jan 4;375(1):217-28. Epub 2007 Oct 17. PMID:18005987[3] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||