2w2m: Difference between revisions
New page: {{Seed}} left|200px <!-- The line below this paragraph, containing "STRUCTURE_2w2m", creates the "Structure Box" on the page. You may change the PDB parameter (which se... |
No edit summary |
||
| (15 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
< | ==WT PCSK9-DELTAC BOUND TO WT EGF-A OF LDLR== | ||
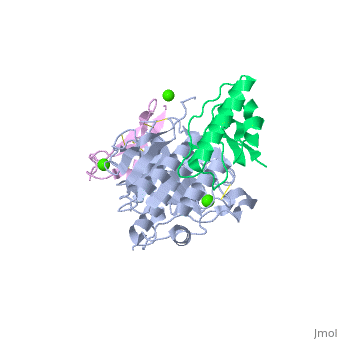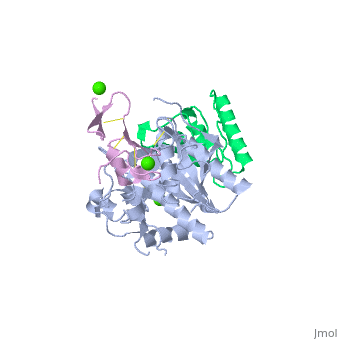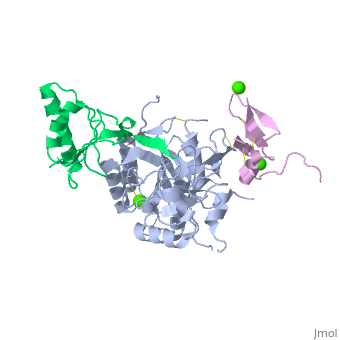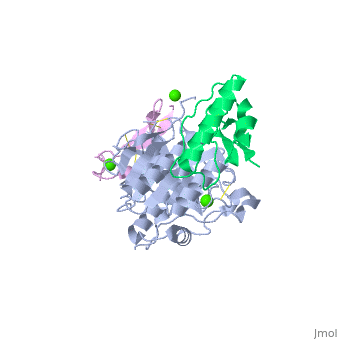
<StructureSection load='2w2m' size='340' side='right'caption='[[2w2m]], [[Resolution|resolution]] 2.40Å' scene=''> | |||
You may | == Structural highlights == | ||
<table><tr><td colspan='2'>[[2w2m]] is a 3 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2W2M OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2W2M FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.4Å</td></tr> | |||
-- | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=CA:CALCIUM+ION'>CA</scene></td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2w2m FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2w2m OCA], [https://pdbe.org/2w2m PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2w2m RCSB], [https://www.ebi.ac.uk/pdbsum/2w2m PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2w2m ProSAT]</span></td></tr> | |||
</table> | |||
== Disease == | |||
[https://www.uniprot.org/uniprot/PCSK9_HUMAN PCSK9_HUMAN] Defects in PCSK9 are the cause of hypercholesterolemia autosomal dominant type 3 (HCHOLA3) [MIM:[https://omim.org/entry/603776 603776]. A familial condition characterized by elevated circulating cholesterol contained in either low-density lipoproteins alone or also in very-low-density lipoproteins.<ref>PMID:12730697</ref> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/PCSK9_HUMAN PCSK9_HUMAN] Crucial player in the regulation of plasma cholesterol homeostasis. Binds to low-density lipid receptor family members: low density lipoprotein receptor (LDLR), very low density lipoprotein receptor (VLDLR), apolipoprotein E receptor (LRP1/APOER) and apolipoprotein receptor 2 (LRP8/APOER2), and promotes their degradation in intracellular acidic compartments. Acts via a non-proteolytic mechanism to enhance the degradation of the hepatic LDLR through a clathrin LDLRAP1/ARH-mediated pathway. May prevent the recycling of LDLR from endosomes to the cell surface or direct it to lysosomes for degradation. Can induce ubiquitination of LDLR leading to its subsequent degradation. Inhibits intracellular degradation of APOB via the autophagosome/lysosome pathway in a LDLR-independent manner. Involved in the disposal of non-acetylated intermediates of BACE1 in the early secretory pathway. Inhibits epithelial Na(+) channel (ENaC)-mediated Na(+) absorption by reducing ENaC surface expression primarily by increasing its proteasomal degradation. Regulates neuronal apoptosis via modulation of LRP8/APOER2 levels and related anti-apoptotic signaling pathways.<ref>PMID:17461796</ref> <ref>PMID:18197702</ref> <ref>PMID:18660751</ref> <ref>PMID:18039658</ref> <ref>PMID:22074827</ref> <ref>PMID:22580899</ref> <ref>PMID:22493497</ref> | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/w2/2w2m_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=2w2m ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
PCSK9 regulates low density lipoprotein receptor (LDLR) levels and consequently is a target for the prevention of atherosclerosis and coronary heart disease. Here we studied the interaction, of LDLR EGF(A/AB) repeats with PCSK9. We show that PCSK9 binds the EGF(AB) repeats in a pH-dependent manner. Although the PCSK9 C-terminal domain is not involved in LDLR binding, PCSK9 autocleavage is required. Moreover, we report the x-ray structure of the PCSK9DeltaC-EGF(AB) complex at neutral pH. Compared with the low pH PCSK9-EGF(A) structure, the new structure revealed rearrangement of the EGF(A) His-306 side chain and disruption of the salt bridge with PCSK9 Asp-374, thus suggesting the basis for enhanced interaction at low pH. In addition, the structure of PCSK9DeltaC bound to EGF(AB)(H306Y), a mutant associated with familial hypercholesterolemia (FH), reveals that the Tyr-306 side chain forms a hydrogen bond with PCSK9 Asp-374, thus mimicking His-306 in the low pH conformation. Consistently, Tyr-306 confers increased affinity for PCSK9. Importantly, we found that although the EGF(AB)(H306Y)-PCSK9 interaction is pH-independent, LDLR(H306Y) binds PCSK9 50-fold better at low pH, suggesting that factors other than His-306 contribute to the pH dependence of PCSK9-LDLR binding. Further, we determined the structures of EGF(AB) bound to PCSK9DeltaC containing the FH-associated D374Y and D374H mutations, revealing additional interactions with EGF(A) mediated by Tyr-374/His-374 and providing a rationale for their disease phenotypes. Finally, we report the inhibitory properties of EGF repeats in a cellular assay measuring LDL uptake. | |||
Structural and biochemical characterization of the wild type PCSK9-EGF(AB) complex and natural familial hypercholesterolemia mutants.,Bottomley MJ, Cirillo A, Orsatti L, Ruggeri L, Fisher TS, Santoro JC, Cummings RT, Cubbon RM, Lo Surdo P, Calzetta A, Noto A, Baysarowich J, Mattu M, Talamo F, De Francesco R, Sparrow CP, Sitlani A, Carfi A J Biol Chem. 2009 Jan 9;284(2):1313-23. Epub 2008 Nov 10. PMID:19001363<ref>PMID:19001363</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 2w2m" style="background-color:#fffaf0;"></div> | |||
== | ==See Also== | ||
*[[LDL receptor|LDL receptor]] | |||
*[[PCSK9|PCSK9]] | |||
== References == | |||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
[[Category: Baysarowich | [[Category: Large Structures]] | ||
[[Category: Bottomley | [[Category: Baysarowich J]] | ||
[[Category: Calzetta | [[Category: Bottomley MJ]] | ||
[[Category: Carfi | [[Category: Calzetta A]] | ||
[[Category: Cirillo | [[Category: Carfi A]] | ||
[[Category: Cubbon | [[Category: Cirillo A]] | ||
[[Category: Cummings | [[Category: Cubbon RM]] | ||
[[Category: Fisher | [[Category: Cummings RT]] | ||
[[Category: | [[Category: De Francesco R]] | ||
[[Category: Mattu | [[Category: Fisher TS]] | ||
[[Category: Noto | [[Category: Lo Surdo P]] | ||
[[Category: Orsatti | [[Category: Mattu M]] | ||
[[Category: Ruggeri | [[Category: Noto A]] | ||
[[Category: Santoro | [[Category: Orsatti L]] | ||
[[Category: Sitlani | [[Category: Ruggeri L]] | ||
[[Category: Sparrow | [[Category: Santoro JC]] | ||
[[Category: Sitlani A]] | |||
[[Category: Talamo | [[Category: Sparrow CP]] | ||
[[Category: Talamo F]] | |||
Latest revision as of 08:33, 17 October 2024
WT PCSK9-DELTAC BOUND TO WT EGF-A OF LDLRWT PCSK9-DELTAC BOUND TO WT EGF-A OF LDLR
Structural highlights
DiseasePCSK9_HUMAN Defects in PCSK9 are the cause of hypercholesterolemia autosomal dominant type 3 (HCHOLA3) [MIM:603776. A familial condition characterized by elevated circulating cholesterol contained in either low-density lipoproteins alone or also in very-low-density lipoproteins.[1] FunctionPCSK9_HUMAN Crucial player in the regulation of plasma cholesterol homeostasis. Binds to low-density lipid receptor family members: low density lipoprotein receptor (LDLR), very low density lipoprotein receptor (VLDLR), apolipoprotein E receptor (LRP1/APOER) and apolipoprotein receptor 2 (LRP8/APOER2), and promotes their degradation in intracellular acidic compartments. Acts via a non-proteolytic mechanism to enhance the degradation of the hepatic LDLR through a clathrin LDLRAP1/ARH-mediated pathway. May prevent the recycling of LDLR from endosomes to the cell surface or direct it to lysosomes for degradation. Can induce ubiquitination of LDLR leading to its subsequent degradation. Inhibits intracellular degradation of APOB via the autophagosome/lysosome pathway in a LDLR-independent manner. Involved in the disposal of non-acetylated intermediates of BACE1 in the early secretory pathway. Inhibits epithelial Na(+) channel (ENaC)-mediated Na(+) absorption by reducing ENaC surface expression primarily by increasing its proteasomal degradation. Regulates neuronal apoptosis via modulation of LRP8/APOER2 levels and related anti-apoptotic signaling pathways.[2] [3] [4] [5] [6] [7] [8] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedPCSK9 regulates low density lipoprotein receptor (LDLR) levels and consequently is a target for the prevention of atherosclerosis and coronary heart disease. Here we studied the interaction, of LDLR EGF(A/AB) repeats with PCSK9. We show that PCSK9 binds the EGF(AB) repeats in a pH-dependent manner. Although the PCSK9 C-terminal domain is not involved in LDLR binding, PCSK9 autocleavage is required. Moreover, we report the x-ray structure of the PCSK9DeltaC-EGF(AB) complex at neutral pH. Compared with the low pH PCSK9-EGF(A) structure, the new structure revealed rearrangement of the EGF(A) His-306 side chain and disruption of the salt bridge with PCSK9 Asp-374, thus suggesting the basis for enhanced interaction at low pH. In addition, the structure of PCSK9DeltaC bound to EGF(AB)(H306Y), a mutant associated with familial hypercholesterolemia (FH), reveals that the Tyr-306 side chain forms a hydrogen bond with PCSK9 Asp-374, thus mimicking His-306 in the low pH conformation. Consistently, Tyr-306 confers increased affinity for PCSK9. Importantly, we found that although the EGF(AB)(H306Y)-PCSK9 interaction is pH-independent, LDLR(H306Y) binds PCSK9 50-fold better at low pH, suggesting that factors other than His-306 contribute to the pH dependence of PCSK9-LDLR binding. Further, we determined the structures of EGF(AB) bound to PCSK9DeltaC containing the FH-associated D374Y and D374H mutations, revealing additional interactions with EGF(A) mediated by Tyr-374/His-374 and providing a rationale for their disease phenotypes. Finally, we report the inhibitory properties of EGF repeats in a cellular assay measuring LDL uptake. Structural and biochemical characterization of the wild type PCSK9-EGF(AB) complex and natural familial hypercholesterolemia mutants.,Bottomley MJ, Cirillo A, Orsatti L, Ruggeri L, Fisher TS, Santoro JC, Cummings RT, Cubbon RM, Lo Surdo P, Calzetta A, Noto A, Baysarowich J, Mattu M, Talamo F, De Francesco R, Sparrow CP, Sitlani A, Carfi A J Biol Chem. 2009 Jan 9;284(2):1313-23. Epub 2008 Nov 10. PMID:19001363[9] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||