1m8v: Difference between revisions
New page: left|200px<br /><applet load="1m8v" size="450" color="white" frame="true" align="right" spinBox="true" caption="1m8v, resolution 2.6Å" /> '''Structure of Pyrococc... |
No edit summary |
||
| (17 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
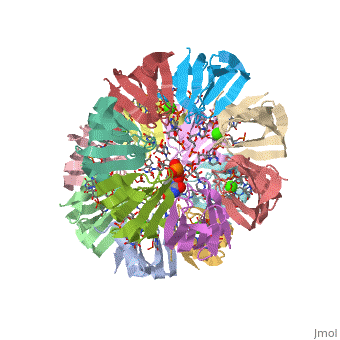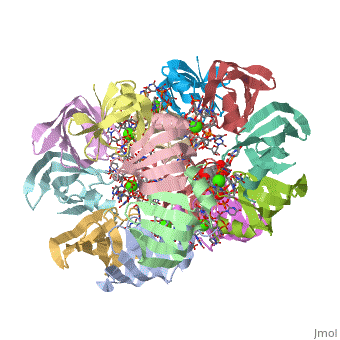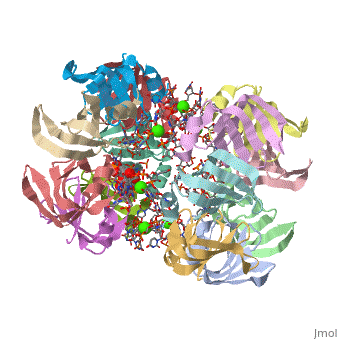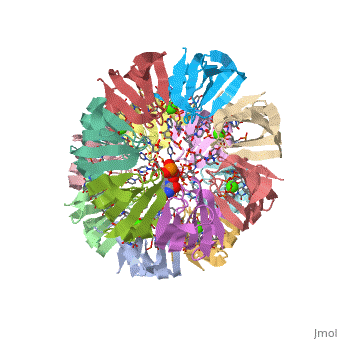
== | ==Structure of Pyrococcus abyssii Sm Protein in Complex with a Uridine Heptamer== | ||
The Sm proteins are conserved in all three domains of life and are always | <StructureSection load='1m8v' size='340' side='right'caption='[[1m8v]], [[Resolution|resolution]] 2.60Å' scene=''> | ||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[1m8v]] is a 21 chain structure with sequence from [https://en.wikipedia.org/wiki/Pyrococcus_abyssi Pyrococcus abyssi]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1M8V OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1M8V FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.6Å</td></tr> | |||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=CA:CALCIUM+ION'>CA</scene>, <scene name='pdbligand=U5P:URIDINE-5-MONOPHOSPHATE'>U5P</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1m8v FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1m8v OCA], [https://pdbe.org/1m8v PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1m8v RCSB], [https://www.ebi.ac.uk/pdbsum/1m8v PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1m8v ProSAT]</span></td></tr> | |||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/RUXX_PYRAB RUXX_PYRAB] | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/m8/1m8v_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1m8v ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
The Sm proteins are conserved in all three domains of life and are always associated with U-rich RNA sequences. Their proposed function is to mediate RNA-RNA interactions. We present here the crystal structures of Pyrococcus abyssi Sm protein (PA-Sm1) and its complex with a uridine heptamer. The overall structure of the protein complex, a heptameric ring with a central cavity, is similar to that proposed for the eukaryotic Sm core complex and found for other archaeal Sm proteins. RNA molecules bind to the protein at two different sites. They interact specifically inside the ring with three highly conserved residues, defining the uridine-binding pocket. In addition, nucleotides also interact on the surface formed by the N-terminal alpha-helix as well as a conserved aromatic residue in beta-strand 2 of the PA-Sm1 protein. The mutation of this conserved aromatic residue shows the importance of this second site for the discrimination between RNA sequences. Given the high structural homology between archaeal and eukaryotic Sm proteins, the PA-Sm1.RNA complex provides a model for how the small nuclear RNA contacts the Sm proteins in the Sm core. In addition, it suggests how Sm proteins might exert their function as modulators of RNA-RNA interactions. | |||
Crystal structures of the Pyrococcus abyssi Sm core and its complex with RNA. Common features of RNA binding in archaea and eukarya.,Thore S, Mayer C, Sauter C, Weeks S, Suck D J Biol Chem. 2003 Jan 10;278(2):1239-47. Epub 2002 Oct 29. PMID:12409299<ref>PMID:12409299</ref> | |||
== | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
</div> | |||
<div class="pdbe-citations 1m8v" style="background-color:#fffaf0;"></div> | |||
==See Also== | |||
*[[Sm-like protein|Sm-like protein]] | |||
*[[Sm-like protein 3D structures|Sm-like protein 3D structures]] | |||
== References == | |||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
[[Category: Large Structures]] | |||
[[Category: Pyrococcus abyssi]] | [[Category: Pyrococcus abyssi]] | ||
[[Category: Mayer C]] | |||
[[Category: Mayer | [[Category: Sauter C]] | ||
[[Category: Sauter | [[Category: Suck D]] | ||
[[Category: Suck | [[Category: Thore S]] | ||
[[Category: Thore | [[Category: Weeks S]] | ||
[[Category: Weeks | |||
Latest revision as of 07:43, 17 October 2024
Structure of Pyrococcus abyssii Sm Protein in Complex with a Uridine HeptamerStructure of Pyrococcus abyssii Sm Protein in Complex with a Uridine Heptamer
Structural highlights
FunctionEvolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedThe Sm proteins are conserved in all three domains of life and are always associated with U-rich RNA sequences. Their proposed function is to mediate RNA-RNA interactions. We present here the crystal structures of Pyrococcus abyssi Sm protein (PA-Sm1) and its complex with a uridine heptamer. The overall structure of the protein complex, a heptameric ring with a central cavity, is similar to that proposed for the eukaryotic Sm core complex and found for other archaeal Sm proteins. RNA molecules bind to the protein at two different sites. They interact specifically inside the ring with three highly conserved residues, defining the uridine-binding pocket. In addition, nucleotides also interact on the surface formed by the N-terminal alpha-helix as well as a conserved aromatic residue in beta-strand 2 of the PA-Sm1 protein. The mutation of this conserved aromatic residue shows the importance of this second site for the discrimination between RNA sequences. Given the high structural homology between archaeal and eukaryotic Sm proteins, the PA-Sm1.RNA complex provides a model for how the small nuclear RNA contacts the Sm proteins in the Sm core. In addition, it suggests how Sm proteins might exert their function as modulators of RNA-RNA interactions. Crystal structures of the Pyrococcus abyssi Sm core and its complex with RNA. Common features of RNA binding in archaea and eukarya.,Thore S, Mayer C, Sauter C, Weeks S, Suck D J Biol Chem. 2003 Jan 10;278(2):1239-47. Epub 2002 Oct 29. PMID:12409299[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||